James Priest
Machine Learning for Automated Mitral Regurgitation Detection from Cardiac Imaging
Oct 07, 2023Abstract:Mitral regurgitation (MR) is a heart valve disease with potentially fatal consequences that can only be forestalled through timely diagnosis and treatment. Traditional diagnosis methods are expensive, labor-intensive and require clinical expertise, posing a barrier to screening for MR. To overcome this impediment, we propose a new semi-supervised model for MR classification called CUSSP. CUSSP operates on cardiac imaging slices of the 4-chamber view of the heart. It uses standard computer vision techniques and contrastive models to learn from large amounts of unlabeled data, in conjunction with specialized classifiers to establish the first ever automated MR classification system. Evaluated on a test set of 179 labeled -- 154 non-MR and 25 MR -- sequences, CUSSP attains an F1 score of 0.69 and a ROC-AUC score of 0.88, setting the first benchmark result for this new task.
* 12 pages including references and the appendix. 9 Figures, 2 tables. Accepted at MICCAI (Machine Learning for Automated Mitral Regurgitation Detection from Cardiac Imaging) 2023, Link to Springer at https://link.springer.com/chapter/10.1007/978-3-031-43990-2_23
Ivy: Instrumental Variable Synthesis for Causal Inference
Apr 11, 2020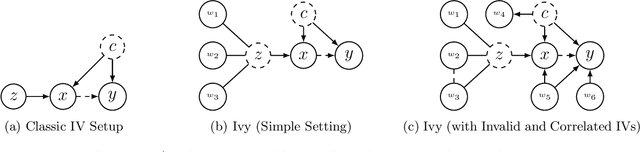
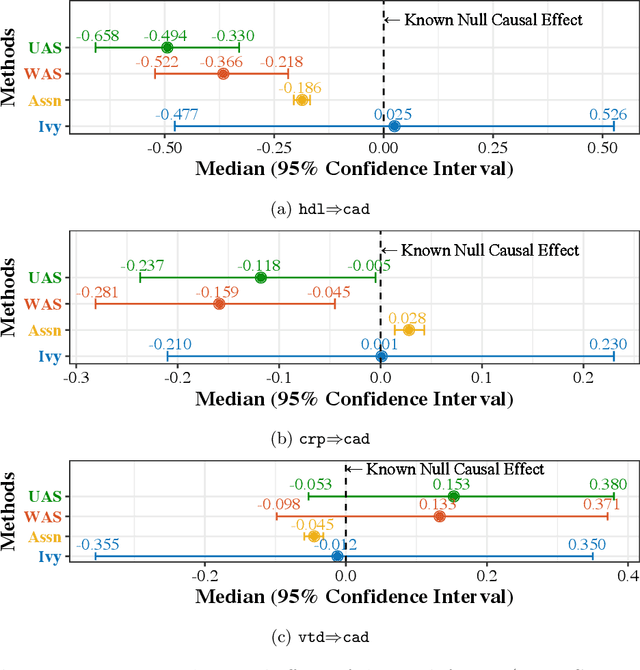
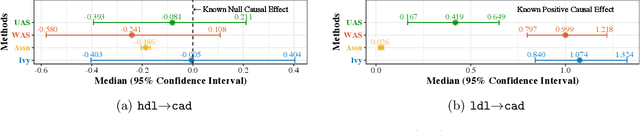
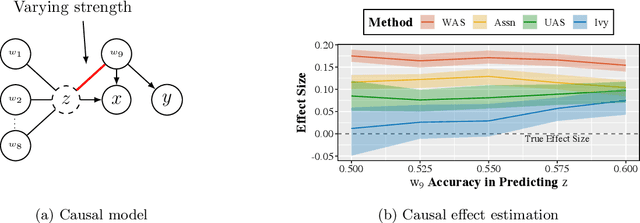
Abstract:A popular way to estimate the causal effect of a variable x on y from observational data is to use an instrumental variable (IV): a third variable z that affects y only through x. The more strongly z is associated with x, the more reliable the estimate is, but such strong IVs are difficult to find. Instead, practitioners combine more commonly available IV candidates---which are not necessarily strong, or even valid, IVs---into a single "summary" that is plugged into causal effect estimators in place of an IV. In genetic epidemiology, such approaches are known as allele scores. Allele scores require strong assumptions---independence and validity of all IV candidates---for the resulting estimate to be reliable. To relax these assumptions, we propose Ivy, a new method to combine IV candidates that can handle correlated and invalid IV candidates in a robust manner. Theoretically, we characterize this robustness, its limits, and its impact on the resulting causal estimates. Empirically, Ivy can correctly identify the directionality of known relationships and is robust against false discovery (median effect size <= 0.025) on three real-world datasets with no causal effects, while allele scores return more biased estimates (median effect size >= 0.118).
Multi-Resolution Weak Supervision for Sequential Data
Oct 21, 2019
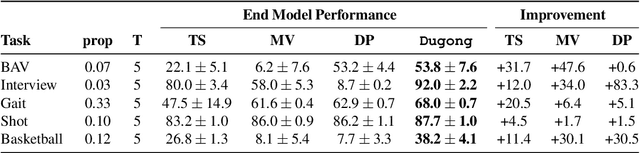
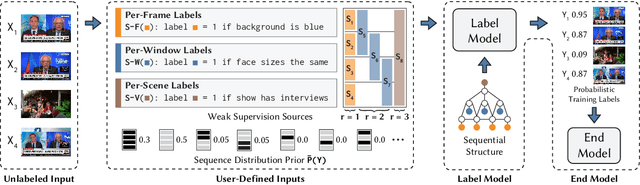

Abstract:Since manually labeling training data is slow and expensive, recent industrial and scientific research efforts have turned to weaker or noisier forms of supervision sources. However, existing weak supervision approaches fail to model multi-resolution sources for sequential data, like video, that can assign labels to individual elements or collections of elements in a sequence. A key challenge in weak supervision is estimating the unknown accuracies and correlations of these sources without using labeled data. Multi-resolution sources exacerbate this challenge due to complex correlations and sample complexity that scales in the length of the sequence. We propose Dugong, the first framework to model multi-resolution weak supervision sources with complex correlations to assign probabilistic labels to training data. Theoretically, we prove that Dugong, under mild conditions, can uniquely recover the unobserved accuracy and correlation parameters and use parameter sharing to improve sample complexity. Our method assigns clinician-validated labels to population-scale biomedical video repositories, helping outperform traditional supervision by 36.8 F1 points and addressing a key use case where machine learning has been severely limited by the lack of expert labeled data. On average, Dugong improves over traditional supervision by 16.0 F1 points and existing weak supervision approaches by 24.2 F1 points across several video and sensor classification tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge