Nimit Sohoni
Llamba: Scaling Distilled Recurrent Models for Efficient Language Processing
Feb 23, 2025Abstract:We introduce Llamba, a family of efficient recurrent language models distilled from Llama-3.x into the Mamba architecture. The series includes Llamba-1B, Llamba-3B, and Llamba-8B, which achieve higher inference throughput and handle significantly larger batch sizes than Transformer-based models while maintaining comparable benchmark performance. Furthermore, Llamba demonstrates the effectiveness of cross-architecture distillation using MOHAWK (Bick et al., 2024), achieving these results with less than 0.1% of the training data typically used for models of similar size. To take full advantage of their efficiency, we provide an optimized implementation of Llamba for resource-constrained devices such as smartphones and edge platforms, offering a practical and memory-efficient alternative to Transformers. Overall, Llamba improves the tradeoff between speed, memory efficiency, and performance, making high-quality language models more accessible.
Monarch: Expressive Structured Matrices for Efficient and Accurate Training
Apr 01, 2022



Abstract:Large neural networks excel in many domains, but they are expensive to train and fine-tune. A popular approach to reduce their compute or memory requirements is to replace dense weight matrices with structured ones (e.g., sparse, low-rank, Fourier transform). These methods have not seen widespread adoption (1) in end-to-end training due to unfavorable efficiency--quality tradeoffs, and (2) in dense-to-sparse fine-tuning due to lack of tractable algorithms to approximate a given dense weight matrix. To address these issues, we propose a class of matrices (Monarch) that is hardware-efficient (they are parameterized as products of two block-diagonal matrices for better hardware utilization) and expressive (they can represent many commonly used transforms). Surprisingly, the problem of approximating a dense weight matrix with a Monarch matrix, though nonconvex, has an analytical optimal solution. These properties of Monarch matrices unlock new ways to train and fine-tune sparse and dense models. We empirically validate that Monarch can achieve favorable accuracy-efficiency tradeoffs in several end-to-end sparse training applications: speeding up ViT and GPT-2 training on ImageNet classification and Wikitext-103 language modeling by 2x with comparable model quality, and reducing the error on PDE solving and MRI reconstruction tasks by 40%. In sparse-to-dense training, with a simple technique called "reverse sparsification," Monarch matrices serve as a useful intermediate representation to speed up GPT-2 pretraining on OpenWebText by 2x without quality drop. The same technique brings 23% faster BERT pretraining than even the very optimized implementation from Nvidia that set the MLPerf 1.1 record. In dense-to-sparse fine-tuning, as a proof-of-concept, our Monarch approximation algorithm speeds up BERT fine-tuning on GLUE by 1.7x with comparable accuracy.
BARACK: Partially Supervised Group Robustness With Guarantees
Dec 31, 2021



Abstract:While neural networks have shown remarkable success on classification tasks in terms of average-case performance, they often fail to perform well on certain groups of the data. Such group information may be expensive to obtain; thus, recent works in robustness and fairness have proposed ways to improve worst-group performance even when group labels are unavailable for the training data. However, these methods generally underperform methods that utilize group information at training time. In this work, we assume access to a small number of group labels alongside a larger dataset without group labels. We propose BARACK, a simple two-step framework to utilize this partial group information to improve worst-group performance: train a model to predict the missing group labels for the training data, and then use these predicted group labels in a robust optimization objective. Theoretically, we provide generalization bounds for our approach in terms of the worst-group performance, showing how the generalization error scales with respect to both the total number of training points and the number of training points with group labels. Empirically, our method outperforms the baselines that do not use group information, even when only 1-33% of points have group labels. We provide ablation studies to support the robustness and extensibility of our framework.
Mandoline: Model Evaluation under Distribution Shift
Jul 01, 2021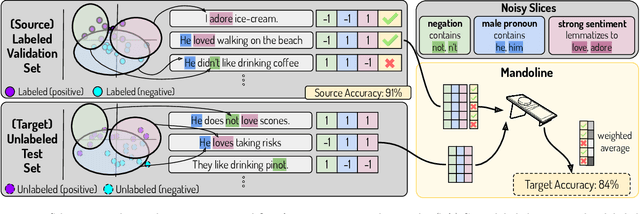
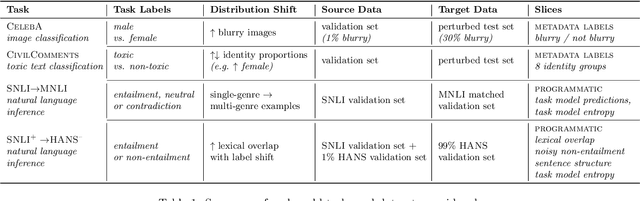
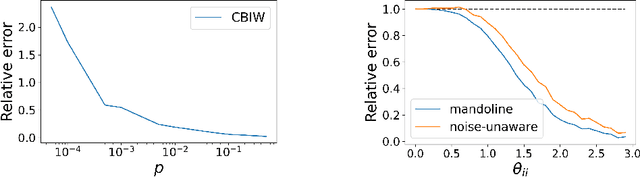
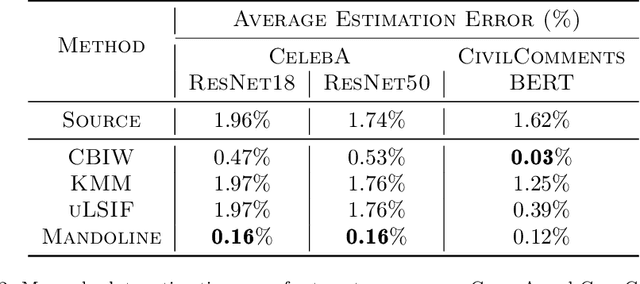
Abstract:Machine learning models are often deployed in different settings than they were trained and validated on, posing a challenge to practitioners who wish to predict how well the deployed model will perform on a target distribution. If an unlabeled sample from the target distribution is available, along with a labeled sample from a possibly different source distribution, standard approaches such as importance weighting can be applied to estimate performance on the target. However, importance weighting struggles when the source and target distributions have non-overlapping support or are high-dimensional. Taking inspiration from fields such as epidemiology and polling, we develop Mandoline, a new evaluation framework that mitigates these issues. Our key insight is that practitioners may have prior knowledge about the ways in which the distribution shifts, which we can use to better guide the importance weighting procedure. Specifically, users write simple "slicing functions" - noisy, potentially correlated binary functions intended to capture possible axes of distribution shift - to compute reweighted performance estimates. We further describe a density ratio estimation framework for the slices and show how its estimation error scales with slice quality and dataset size. Empirical validation on NLP and vision tasks shows that \name can estimate performance on the target distribution up to $3\times$ more accurately compared to standard baselines.
Ivy: Instrumental Variable Synthesis for Causal Inference
Apr 11, 2020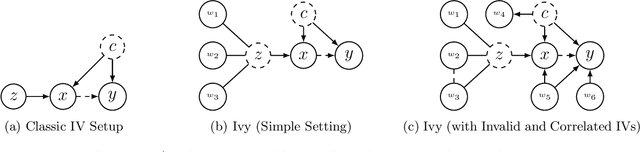
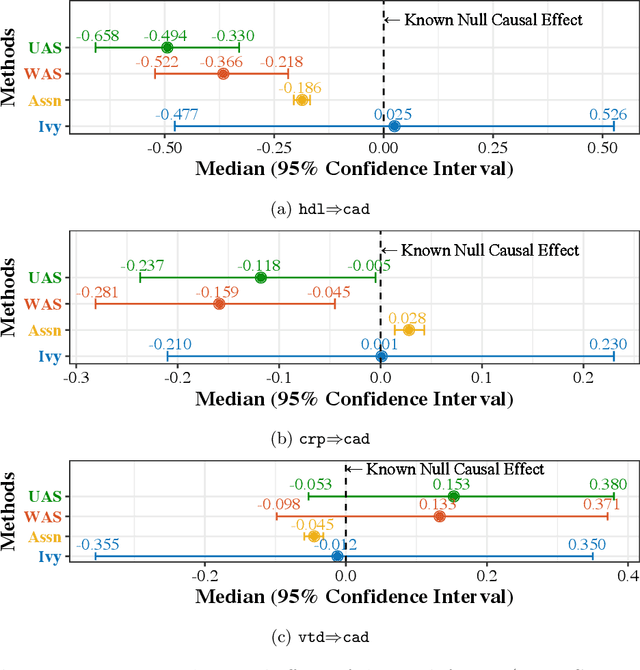
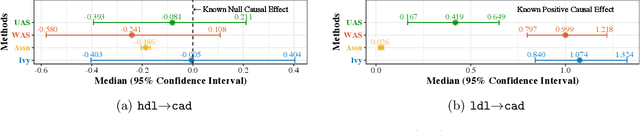
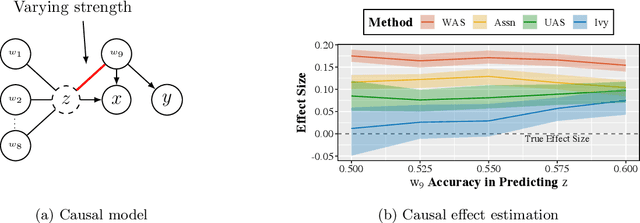
Abstract:A popular way to estimate the causal effect of a variable x on y from observational data is to use an instrumental variable (IV): a third variable z that affects y only through x. The more strongly z is associated with x, the more reliable the estimate is, but such strong IVs are difficult to find. Instead, practitioners combine more commonly available IV candidates---which are not necessarily strong, or even valid, IVs---into a single "summary" that is plugged into causal effect estimators in place of an IV. In genetic epidemiology, such approaches are known as allele scores. Allele scores require strong assumptions---independence and validity of all IV candidates---for the resulting estimate to be reliable. To relax these assumptions, we propose Ivy, a new method to combine IV candidates that can handle correlated and invalid IV candidates in a robust manner. Theoretically, we characterize this robustness, its limits, and its impact on the resulting causal estimates. Empirically, Ivy can correctly identify the directionality of known relationships and is robust against false discovery (median effect size <= 0.025) on three real-world datasets with no causal effects, while allele scores return more biased estimates (median effect size >= 0.118).
DeepHeart: Semi-Supervised Sequence Learning for Cardiovascular Risk Prediction
Feb 07, 2018



Abstract:We train and validate a semi-supervised, multi-task LSTM on 57,675 person-weeks of data from off-the-shelf wearable heart rate sensors, showing high accuracy at detecting multiple medical conditions, including diabetes (0.8451), high cholesterol (0.7441), high blood pressure (0.8086), and sleep apnea (0.8298). We compare two semi-supervised train- ing methods, semi-supervised sequence learning and heuristic pretraining, and show they outperform hand-engineered biomarkers from the medical literature. We believe our work suggests a new approach to patient risk stratification based on cardiovascular risk scores derived from popular wearables such as Fitbit, Apple Watch, or Android Wear.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge