Geoffrey H. Tison
BenchMD: A Benchmark for Modality-Agnostic Learning on Medical Images and Sensors
Apr 17, 2023



Abstract:Medical data poses a daunting challenge for AI algorithms: it exists in many different modalities, experiences frequent distribution shifts, and suffers from a scarcity of examples and labels. Recent advances, including transformers and self-supervised learning, promise a more universal approach that can be applied flexibly across these diverse conditions. To measure and drive progress in this direction, we present BenchMD: a benchmark that tests how modality-agnostic methods, including architectures and training techniques (e.g. self-supervised learning, ImageNet pretraining), perform on a diverse array of clinically-relevant medical tasks. BenchMD combines 19 publicly available datasets for 7 medical modalities, including 1D sensor data, 2D images, and 3D volumetric scans. Our benchmark reflects real-world data constraints by evaluating methods across a range of dataset sizes, including challenging few-shot settings that incentivize the use of pretraining. Finally, we evaluate performance on out-of-distribution data collected at different hospitals than the training data, representing naturally-occurring distribution shifts that frequently degrade the performance of medical AI models. Our baseline results demonstrate that no modality-agnostic technique achieves strong performance across all modalities, leaving ample room for improvement on the benchmark. Code is released at https://github.com/rajpurkarlab/BenchMD .
CathAI: Fully Automated Interpretation of Coronary Angiograms Using Neural Networks
Jun 14, 2021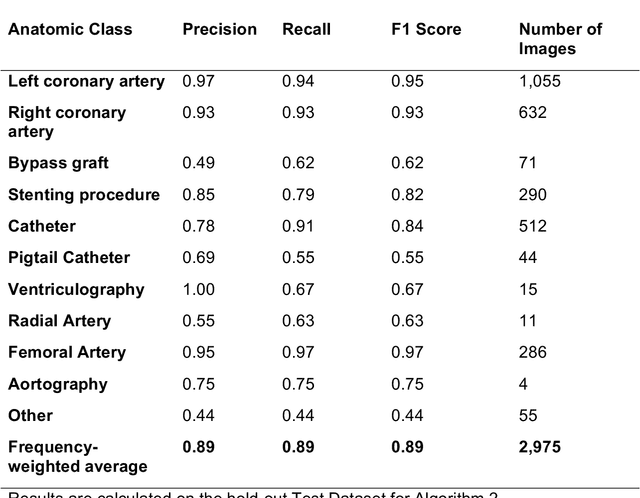
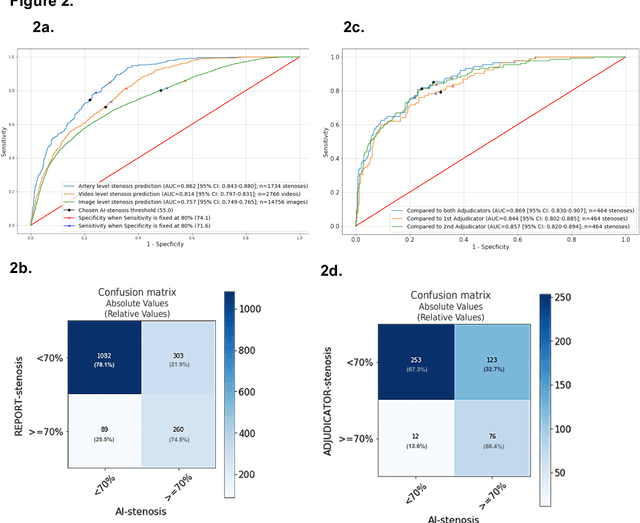
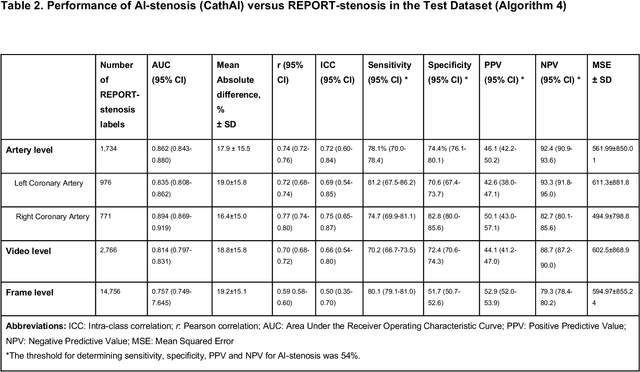
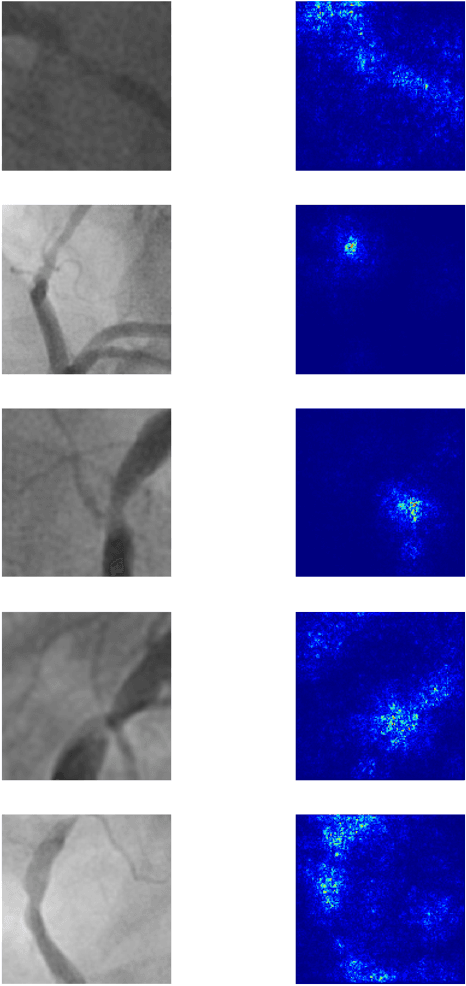
Abstract:Coronary heart disease (CHD) is the leading cause of adult death in the United States and worldwide, and for which the coronary angiography procedure is the primary gateway for diagnosis and clinical management decisions. The standard-of-care for interpretation of coronary angiograms depends upon ad-hoc visual assessment by the physician operator. However, ad-hoc visual interpretation of angiograms is poorly reproducible, highly variable and bias prone. Here we show for the first time that fully-automated angiogram interpretation to estimate coronary artery stenosis is possible using a sequence of deep neural network algorithms. The algorithmic pipeline we developed--called CathAI--achieves state-of-the art performance across the sequence of tasks required to accomplish automated interpretation of unselected, real-world angiograms. CathAI (Algorithms 1-2) demonstrated positive predictive value, sensitivity and F1 score of >=90% to identify the projection angle overall and >=93% for left or right coronary artery angiogram detection, the primary anatomic structures of interest. To predict obstructive coronary artery stenosis (>=70% stenosis), CathAI (Algorithm 4) exhibited an area under the receiver operating characteristic curve (AUC) of 0.862 (95% CI: 0.843-0.880). When externally validated in a healthcare system in another country, CathAI AUC was 0.869 (95% CI: 0.830-0.907) to predict obstructive coronary artery stenosis. Our results demonstrate that multiple purpose-built neural networks can function in sequence to accomplish the complex series of tasks required for automated analysis of real-world angiograms. Deployment of CathAI may serve to increase standardization and reproducibility in coronary stenosis assessment, while providing a robust foundation to accomplish future tasks for algorithmic angiographic interpretation.
Using Multitask Learning to Improve 12-Lead Electrocardiogram Classification
Dec 04, 2018


Abstract:We develop a multi-task convolutional neural network (CNN) to classify multiple diagnoses from 12-lead electrocardiograms (ECGs) using a dataset comprised of over 40,000 ECGs, with labels derived from cardiologist clinical interpretations. Since many clinically important classes can occur in low frequencies, approaches are needed to improve performance on rare classes. We compare the performance of several single-class classifiers on rare classes to a multi-headed classifier across all available classes. We demonstrate that the addition of common classes can significantly improve CNN performance on rarer classes when compared to a model trained on the rarer class in isolation. Using this method, we develop a model with high performance as measured by F1 score on multiple clinically relevant classes compared against the gold-standard cardiologist interpretation.
Automated and Interpretable Patient ECG Profiles for Disease Detection, Tracking, and Discovery
Jul 06, 2018



Abstract:The electrocardiogram or ECG has been in use for over 100 years and remains the most widely performed diagnostic test to characterize cardiac structure and electrical activity. We hypothesized that parallel advances in computing power, innovations in machine learning algorithms, and availability of large-scale digitized ECG data would enable extending the utility of the ECG beyond its current limitations, while at the same time preserving interpretability, which is fundamental to medical decision-making. We identified 36,186 ECGs from the UCSF database that were 1) in normal sinus rhythm and 2) would enable training of specific models for estimation of cardiac structure or function or detection of disease. We derived a novel model for ECG segmentation using convolutional neural networks (CNN) and Hidden Markov Models (HMM) and evaluated its output by comparing electrical interval estimates to 141,864 measurements from the clinical workflow. We built a 725-element patient-level ECG profile using downsampled segmentation data and trained machine learning models to estimate left ventricular mass, left atrial volume, mitral annulus e' and to detect and track four diseases: pulmonary arterial hypertension (PAH), hypertrophic cardiomyopathy (HCM), cardiac amyloid (CA), and mitral valve prolapse (MVP). CNN-HMM derived ECG segmentation agreed with clinical estimates, with median absolute deviations (MAD) as a fraction of observed value of 0.6% for heart rate and 4% for QT interval. Patient-level ECG profiles enabled quantitative estimates of left ventricular and mitral annulus e' velocity with good discrimination in binary classification models of left ventricular hypertrophy and diastolic function. Models for disease detection ranged from AUROC of 0.94 to 0.77 for MVP. Top-ranked variables for all models included known ECG characteristics along with novel predictors of these traits/diseases.
DeepHeart: Semi-Supervised Sequence Learning for Cardiovascular Risk Prediction
Feb 07, 2018



Abstract:We train and validate a semi-supervised, multi-task LSTM on 57,675 person-weeks of data from off-the-shelf wearable heart rate sensors, showing high accuracy at detecting multiple medical conditions, including diabetes (0.8451), high cholesterol (0.7441), high blood pressure (0.8086), and sleep apnea (0.8298). We compare two semi-supervised train- ing methods, semi-supervised sequence learning and heuristic pretraining, and show they outperform hand-engineered biomarkers from the medical literature. We believe our work suggests a new approach to patient risk stratification based on cardiovascular risk scores derived from popular wearables such as Fitbit, Apple Watch, or Android Wear.
A Computer Vision Pipeline for Automated Determination of Cardiac Structure and Function and Detection of Disease by Two-Dimensional Echocardiography
Jan 12, 2018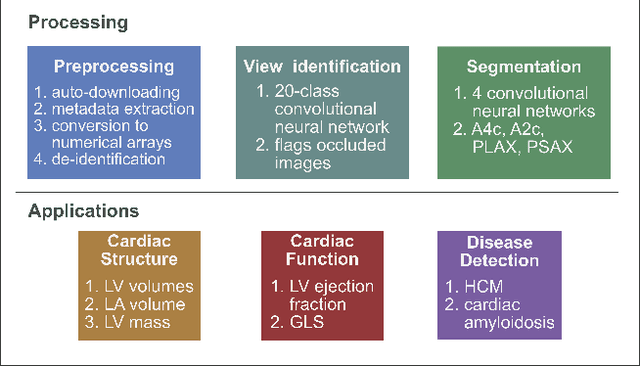
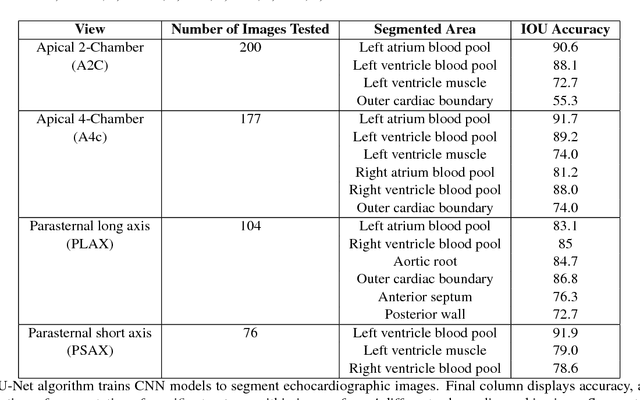
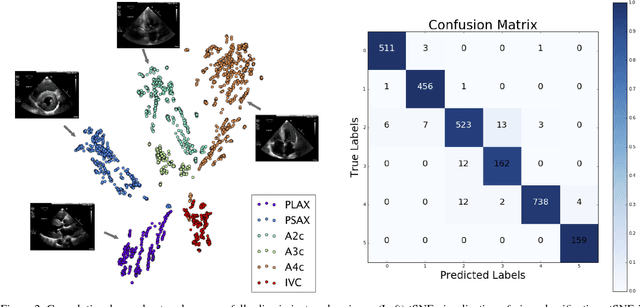
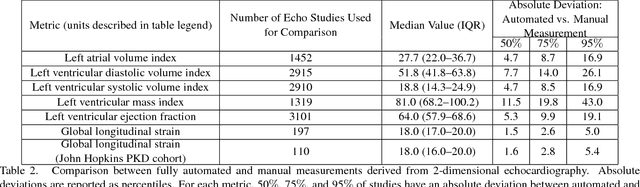
Abstract:Automated cardiac image interpretation has the potential to transform clinical practice in multiple ways including enabling low-cost serial assessment of cardiac function in the primary care and rural setting. We hypothesized that advances in computer vision could enable building a fully automated, scalable analysis pipeline for echocardiogram (echo) interpretation. Our approach entailed: 1) preprocessing; 2) convolutional neural networks (CNN) for view identification, image segmentation, and phasing of the cardiac cycle; 3) quantification of chamber volumes and left ventricular mass; 4) particle tracking to compute longitudinal strain; and 5) targeted disease detection. CNNs accurately identified views (e.g. 99% for apical 4-chamber) and segmented individual cardiac chambers. Cardiac structure measurements agreed with study report values (e.g. mean absolute deviations (MAD) of 7.7 mL/kg/m2 for left ventricular diastolic volume index, 2918 studies). We computed automated ejection fraction and longitudinal strain measurements (within 2 cohorts), which agreed with commercial software-derived values [for ejection fraction, MAD=5.3%, N=3101 studies; for strain, MAD=1.5% (n=197) and 1.6% (n=110)], and demonstrated applicability to serial monitoring of breast cancer patients for trastuzumab cardiotoxicity. Overall, we found that, compared to manual measurements, automated measurements had superior performance across seven internal consistency metrics with an average increase in the Spearman correlation coefficient of 0.05 (p=0.02). Finally, we developed disease detection algorithms for hypertrophic cardiomyopathy and cardiac amyloidosis, with C-statistics of 0.93 and 0.84, respectively. Our pipeline lays the groundwork for using automated interpretation to support point-of-care handheld cardiac ultrasound and large-scale analysis of the millions of echos archived within healthcare systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge