Arjun Desai
Llamba: Scaling Distilled Recurrent Models for Efficient Language Processing
Feb 23, 2025Abstract:We introduce Llamba, a family of efficient recurrent language models distilled from Llama-3.x into the Mamba architecture. The series includes Llamba-1B, Llamba-3B, and Llamba-8B, which achieve higher inference throughput and handle significantly larger batch sizes than Transformer-based models while maintaining comparable benchmark performance. Furthermore, Llamba demonstrates the effectiveness of cross-architecture distillation using MOHAWK (Bick et al., 2024), achieving these results with less than 0.1% of the training data typically used for models of similar size. To take full advantage of their efficiency, we provide an optimized implementation of Llamba for resource-constrained devices such as smartphones and edge platforms, offering a practical and memory-efficient alternative to Transformers. Overall, Llamba improves the tradeoff between speed, memory efficiency, and performance, making high-quality language models more accessible.
Prospector Heads: Generalized Feature Attribution for Large Models & Data
Feb 18, 2024Abstract:Feature attribution, the ability to localize regions of the input data that are relevant for classification, is an important capability for machine learning models in scientific and biomedical domains. Current methods for feature attribution, which rely on "explaining" the predictions of end-to-end classifiers, suffer from imprecise feature localization and are inadequate for use with small sample sizes and high-dimensional datasets due to computational challenges. We introduce prospector heads, an efficient and interpretable alternative to explanation-based methods for feature attribution that can be applied to any encoder and any data modality. Prospector heads generalize across modalities through experiments on sequences (text), images (pathology), and graphs (protein structures), outperforming baseline attribution methods by up to 49 points in mean localization AUPRC. We also demonstrate how prospector heads enable improved interpretation and discovery of class-specific patterns in the input data. Through their high performance, flexibility, and generalizability, prospectors provide a framework for improving trust and transparency for machine learning models in complex domains.
Deep Anatomical Federated Network (Dafne): an open client/server framework for the continuous collaborative improvement of deep-learning-based medical image segmentation
Feb 14, 2023Abstract:Semantic segmentation is a crucial step to extract quantitative information from medical (and, specifically, radiological) images to aid the diagnostic process, clinical follow-up. and to generate biomarkers for clinical research. In recent years, machine learning algorithms have become the primary tool for this task. However, its real-world performance is heavily reliant on the comprehensiveness of training data. Dafne is the first decentralized, collaborative solution that implements continuously evolving deep learning models exploiting the collective knowledge of the users of the system. In the Dafne workflow, the result of each automated segmentation is refined by the user through an integrated interface, so that the new information is used to continuously expand the training pool via federated incremental learning. The models deployed through Dafne are able to improve their performance over time and to generalize to data types not seen in the training sets, thus becoming a viable and practical solution for real-life medical segmentation tasks.
Comp2Comp: Open-Source Body Composition Assessment on Computed Tomography
Feb 13, 2023Abstract:Computed tomography (CT) is routinely used in clinical practice to evaluate a wide variety of medical conditions. While CT scans provide diagnoses, they also offer the ability to extract quantitative body composition metrics to analyze tissue volume and quality. Extracting quantitative body composition measures manually from CT scans is a cumbersome and time-consuming task. Proprietary software has been developed recently to automate this process, but the closed-source nature impedes widespread use. There is a growing need for fully automated body composition software that is more accessible and easier to use, especially for clinicians and researchers who are not experts in medical image processing. To this end, we have built Comp2Comp, an open-source Python package for rapid and automated body composition analysis of CT scans. This package offers models, post-processing heuristics, body composition metrics, automated batching, and polychromatic visualizations. Comp2Comp currently computes body composition measures for bone, skeletal muscle, visceral adipose tissue, and subcutaneous adipose tissue on CT scans of the abdomen. We have created two pipelines for this purpose. The first pipeline computes vertebral measures, as well as muscle and adipose tissue measures, at the T12 - L5 vertebral levels from abdominal CT scans. The second pipeline computes muscle and adipose tissue measures on user-specified 2D axial slices. In this guide, we discuss the architecture of the Comp2Comp pipelines, provide usage instructions, and report internal and external validation results to measure the quality of segmentations and body composition measures. Comp2Comp can be found at https://github.com/StanfordMIMI/Comp2Comp.
Scale-Agnostic Super-Resolution in MRI using Feature-Based Coordinate Networks
Oct 18, 2022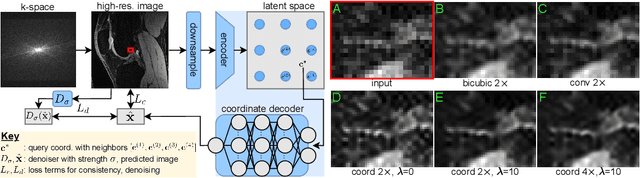
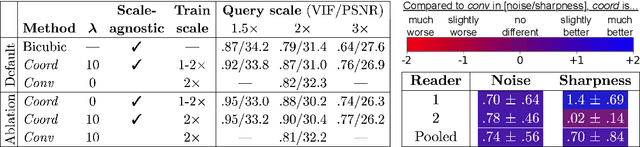
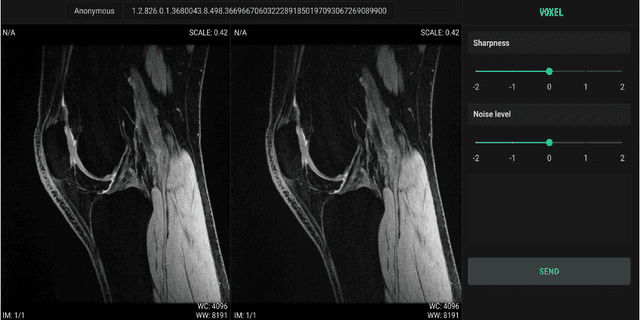
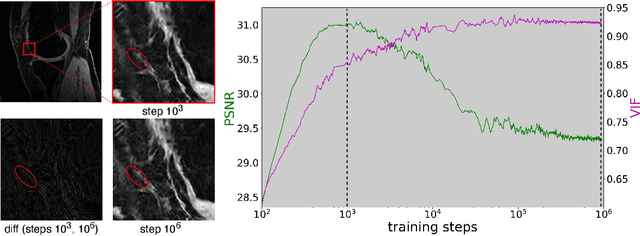
Abstract:We propose using a coordinate network decoder for the task of super-resolution in MRI. The continuous signal representation of coordinate networks enables this approach to be scale-agnostic, i.e. one can train over a continuous range of scales and subsequently query at arbitrary resolutions. Due to the difficulty of performing super-resolution on inherently noisy data, we analyze network behavior under multiple denoising strategies. Lastly we compare this method to a standard convolutional decoder using both quantitative metrics and a radiologist study implemented in Voxel, our newly developed tool for web-based evaluation of medical images.
Monarch: Expressive Structured Matrices for Efficient and Accurate Training
Apr 01, 2022



Abstract:Large neural networks excel in many domains, but they are expensive to train and fine-tune. A popular approach to reduce their compute or memory requirements is to replace dense weight matrices with structured ones (e.g., sparse, low-rank, Fourier transform). These methods have not seen widespread adoption (1) in end-to-end training due to unfavorable efficiency--quality tradeoffs, and (2) in dense-to-sparse fine-tuning due to lack of tractable algorithms to approximate a given dense weight matrix. To address these issues, we propose a class of matrices (Monarch) that is hardware-efficient (they are parameterized as products of two block-diagonal matrices for better hardware utilization) and expressive (they can represent many commonly used transforms). Surprisingly, the problem of approximating a dense weight matrix with a Monarch matrix, though nonconvex, has an analytical optimal solution. These properties of Monarch matrices unlock new ways to train and fine-tune sparse and dense models. We empirically validate that Monarch can achieve favorable accuracy-efficiency tradeoffs in several end-to-end sparse training applications: speeding up ViT and GPT-2 training on ImageNet classification and Wikitext-103 language modeling by 2x with comparable model quality, and reducing the error on PDE solving and MRI reconstruction tasks by 40%. In sparse-to-dense training, with a simple technique called "reverse sparsification," Monarch matrices serve as a useful intermediate representation to speed up GPT-2 pretraining on OpenWebText by 2x without quality drop. The same technique brings 23% faster BERT pretraining than even the very optimized implementation from Nvidia that set the MLPerf 1.1 record. In dense-to-sparse fine-tuning, as a proof-of-concept, our Monarch approximation algorithm speeds up BERT fine-tuning on GLUE by 1.7x with comparable accuracy.
Using Computer Vision to Automate Hand Detection and Tracking of Surgeon Movements in Videos of Open Surgery
Dec 13, 2020



Abstract:Open, or non-laparoscopic surgery, represents the vast majority of all operating room procedures, but few tools exist to objectively evaluate these techniques at scale. Current efforts involve human expert-based visual assessment. We leverage advances in computer vision to introduce an automated approach to video analysis of surgical execution. A state-of-the-art convolutional neural network architecture for object detection was used to detect operating hands in open surgery videos. Automated assessment was expanded by combining model predictions with a fast object tracker to enable surgeon-specific hand tracking. To train our model, we used publicly available videos of open surgery from YouTube and annotated these with spatial bounding boxes of operating hands. Our model's spatial detections of operating hands significantly outperforms the detections achieved using pre-existing hand-detection datasets, and allow for insights into intra-operative movement patterns and economy of motion.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge