Rogier van der Sluijs
MedVAE: Efficient Automated Interpretation of Medical Images with Large-Scale Generalizable Autoencoders
Feb 20, 2025



Abstract:Medical images are acquired at high resolutions with large fields of view in order to capture fine-grained features necessary for clinical decision-making. Consequently, training deep learning models on medical images can incur large computational costs. In this work, we address the challenge of downsizing medical images in order to improve downstream computational efficiency while preserving clinically-relevant features. We introduce MedVAE, a family of six large-scale 2D and 3D autoencoders capable of encoding medical images as downsized latent representations and decoding latent representations back to high-resolution images. We train MedVAE autoencoders using a novel two-stage training approach with 1,052,730 medical images. Across diverse tasks obtained from 20 medical image datasets, we demonstrate that (1) utilizing MedVAE latent representations in place of high-resolution images when training downstream models can lead to efficiency benefits (up to 70x improvement in throughput) while simultaneously preserving clinically-relevant features and (2) MedVAE can decode latent representations back to high-resolution images with high fidelity. Our work demonstrates that large-scale, generalizable autoencoders can help address critical efficiency challenges in the medical domain. Our code is available at https://github.com/StanfordMIMI/MedVAE.
Exploring Image Augmentations for Siamese Representation Learning with Chest X-Rays
Jan 30, 2023



Abstract:Image augmentations are quintessential for effective visual representation learning across self-supervised learning techniques. While augmentation strategies for natural imaging have been studied extensively, medical images are vastly different from their natural counterparts. Thus, it is unknown whether common augmentation strategies employed in Siamese representation learning generalize to medical images and to what extent. To address this challenge, in this study, we systematically assess the effect of various augmentations on the quality and robustness of the learned representations. We train and evaluate Siamese Networks for abnormality detection on chest X-Rays across three large datasets (MIMIC-CXR, CheXpert and VinDR-CXR). We investigate the efficacy of the learned representations through experiments involving linear probing, fine-tuning, zero-shot transfer, and data efficiency. Finally, we identify a set of augmentations that yield robust representations that generalize well to both out-of-distribution data and diseases, while outperforming supervised baselines using just zero-shot transfer and linear probes by up to 20%.
Scale-Agnostic Super-Resolution in MRI using Feature-Based Coordinate Networks
Oct 18, 2022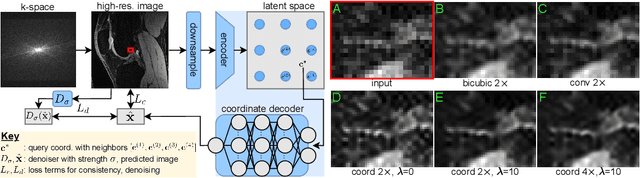
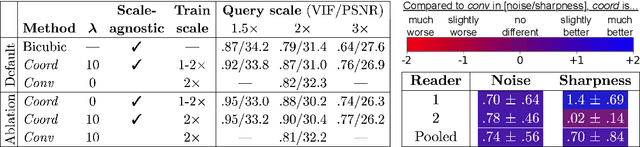
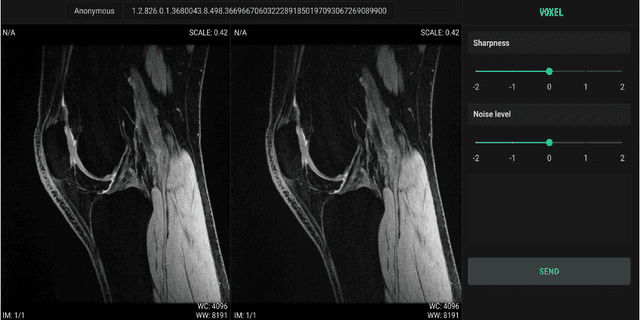
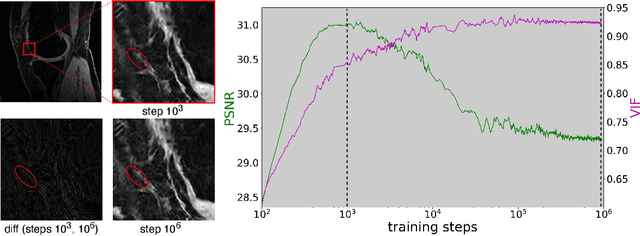
Abstract:We propose using a coordinate network decoder for the task of super-resolution in MRI. The continuous signal representation of coordinate networks enables this approach to be scale-agnostic, i.e. one can train over a continuous range of scales and subsequently query at arbitrary resolutions. Due to the difficulty of performing super-resolution on inherently noisy data, we analyze network behavior under multiple denoising strategies. Lastly we compare this method to a standard convolutional decoder using both quantitative metrics and a radiologist study implemented in Voxel, our newly developed tool for web-based evaluation of medical images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge