Saelig Khattar
Trove: Ontology-driven weak supervision for medical entity classification
Aug 05, 2020



Abstract:Motivation: Recognizing named entities (NER) and their associated attributes like negation are core tasks in natural language processing. However, manually labeling data for entity tasks is time consuming and expensive, creating barriers to using machine learning in new medical applications. Weakly supervised learning, which automatically builds imperfect training sets from low cost, less accurate labeling rules, offers a potential solution. Medical ontologies are compelling sources for generating labels, however combining multiple ontologies without ground truth data creates challenges due to label noise introduced by conflicting entity definitions. Key questions remain on the extent to which weakly supervised entity classification can be automated using ontologies, or how much additional task-specific rule engineering is required for state-of-the-art performance. Also unclear is how pre-trained language models, such as BioBERT, improve the ability to generalize from imperfectly labeled data. Results: We present Trove, a framework for weakly supervised entity classification using medical ontologies. We report state-of-the-art, weakly supervised performance on two NER benchmark datasets and establish new baselines for two entity classification tasks in clinical text. We perform within an average of 3.5 F1 points (4.2%) of NER classifiers trained with hand-labeled data. Automatically learning label source accuracies to correct for label noise provided an average improvement of 3.9 F1 points. BioBERT provided an average improvement of 0.9 F1 points. We measure the impact of combining large numbers of ontologies and present a case study on rapidly building classifiers for COVID-19 clinical tasks. Our framework demonstrates how a wide range of medical entity classifiers can be quickly constructed using weak supervision and without requiring manually-labeled training data.
Multi-Resolution Weak Supervision for Sequential Data
Oct 21, 2019
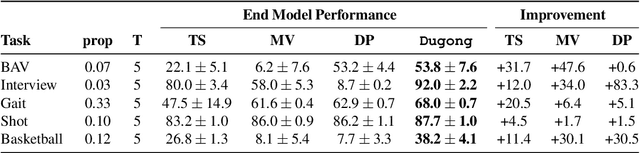
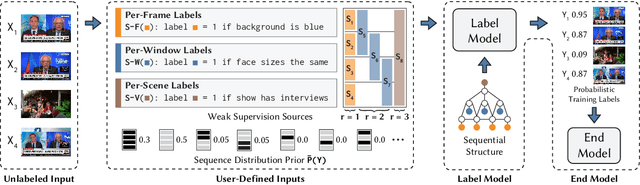

Abstract:Since manually labeling training data is slow and expensive, recent industrial and scientific research efforts have turned to weaker or noisier forms of supervision sources. However, existing weak supervision approaches fail to model multi-resolution sources for sequential data, like video, that can assign labels to individual elements or collections of elements in a sequence. A key challenge in weak supervision is estimating the unknown accuracies and correlations of these sources without using labeled data. Multi-resolution sources exacerbate this challenge due to complex correlations and sample complexity that scales in the length of the sequence. We propose Dugong, the first framework to model multi-resolution weak supervision sources with complex correlations to assign probabilistic labels to training data. Theoretically, we prove that Dugong, under mild conditions, can uniquely recover the unobserved accuracy and correlation parameters and use parameter sharing to improve sample complexity. Our method assigns clinician-validated labels to population-scale biomedical video repositories, helping outperform traditional supervision by 36.8 F1 points and addressing a key use case where machine learning has been severely limited by the lack of expert labeled data. On average, Dugong improves over traditional supervision by 16.0 F1 points and existing weak supervision approaches by 24.2 F1 points across several video and sensor classification tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge