Madalina Fiterau
Audio-Visual Speech Separation via Bottleneck Iterative Network
Jul 09, 2025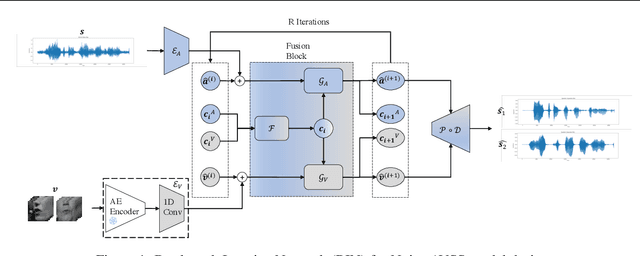
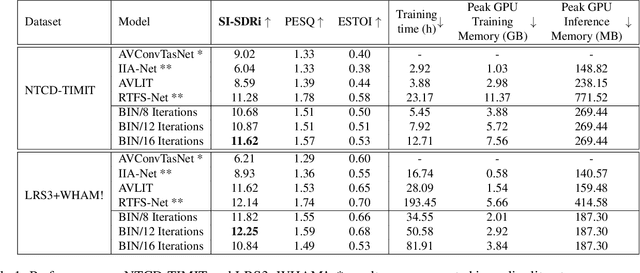
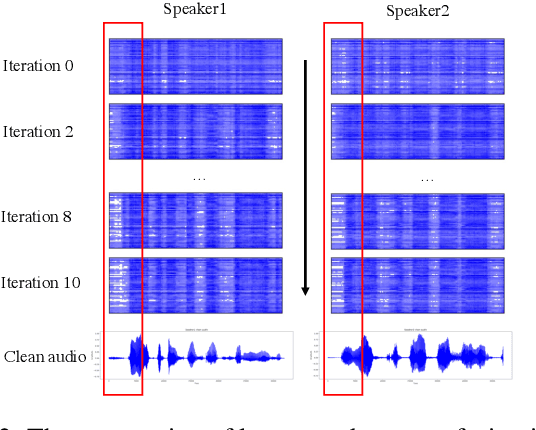
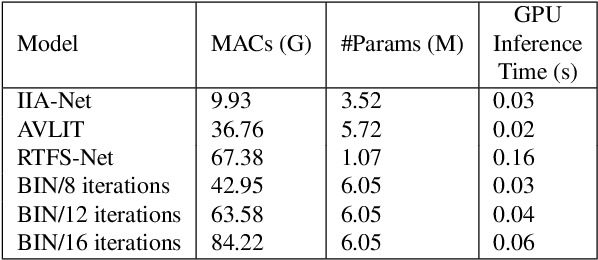
Abstract:Integration of information from non-auditory cues can significantly improve the performance of speech-separation models. Often such models use deep modality-specific networks to obtain unimodal features, and risk being too costly or lightweight but lacking capacity. In this work, we present an iterative representation refinement approach called Bottleneck Iterative Network (BIN), a technique that repeatedly progresses through a lightweight fusion block, while bottlenecking fusion representations by fusion tokens. This helps improve the capacity of the model, while avoiding major increase in model size and balancing between the model performance and training cost. We test BIN on challenging noisy audio-visual speech separation tasks, and show that our approach consistently outperforms state-of-the-art benchmark models with respect to SI-SDRi on NTCD-TIMIT and LRS3+WHAM! datasets, while simultaneously achieving a reduction of more than 50% in training and GPU inference time across nearly all settings.
Leveraging Foundation Language Models (FLMs) for Automated Cohort Extraction from Large EHR Databases
Dec 16, 2024Abstract:A crucial step in cohort studies is to extract the required cohort from one or more study datasets. This step is time-consuming, especially when a researcher is presented with a dataset that they have not previously worked with. When the cohort has to be extracted from multiple datasets, cohort extraction can be extremely laborious. In this study, we present an approach for partially automating cohort extraction from multiple electronic health record (EHR) databases. We formulate the guided multi-dataset cohort extraction problem in which selection criteria are first converted into queries, translating them from natural language text to language that maps to database entities. Then, using FLMs, columns of interest identified from the queries are automatically matched between the study databases. Finally, the generated queries are run across all databases to extract the study cohort. We propose and evaluate an algorithm for automating column matching on two large, popular and publicly-accessible EHR databases -- MIMIC-III and eICU. Our approach achieves a high top-three accuracy of $92\%$, correctly matching $12$ out of the $13$ columns of interest, when using a small, pre-trained general purpose language model. Furthermore, this accuracy is maintained even as the search space (i.e., size of the database) increases.
A/B testing under Interference with Partial Network Information
Apr 16, 2024



Abstract:A/B tests are often required to be conducted on subjects that might have social connections. For e.g., experiments on social media, or medical and social interventions to control the spread of an epidemic. In such settings, the SUTVA assumption for randomized-controlled trials is violated due to network interference, or spill-over effects, as treatments to group A can potentially also affect the control group B. When the underlying social network is known exactly, prior works have demonstrated how to conduct A/B tests adequately to estimate the global average treatment effect (GATE). However, in practice, it is often impossible to obtain knowledge about the exact underlying network. In this paper, we present UNITE: a novel estimator that relax this assumption and can identify GATE while only relying on knowledge of the superset of neighbors for any subject in the graph. Through theoretical analysis and extensive experiments, we show that the proposed approach performs better in comparison to standard estimators.
Zero-shot Microclimate Prediction with Deep Learning
Jan 05, 2024Abstract:Weather station data is a valuable resource for climate prediction, however, its reliability can be limited in remote locations. To compound the issue, making local predictions often relies on sensor data that may not be accessible for a new, previously unmonitored location. In response to these challenges, we propose a novel zero-shot learning approach designed to forecast various climate measurements at new and unmonitored locations. Our method surpasses conventional weather forecasting techniques in predicting microclimate variables by leveraging knowledge extracted from other geographic locations.
Machine Learning for Automated Mitral Regurgitation Detection from Cardiac Imaging
Oct 07, 2023Abstract:Mitral regurgitation (MR) is a heart valve disease with potentially fatal consequences that can only be forestalled through timely diagnosis and treatment. Traditional diagnosis methods are expensive, labor-intensive and require clinical expertise, posing a barrier to screening for MR. To overcome this impediment, we propose a new semi-supervised model for MR classification called CUSSP. CUSSP operates on cardiac imaging slices of the 4-chamber view of the heart. It uses standard computer vision techniques and contrastive models to learn from large amounts of unlabeled data, in conjunction with specialized classifiers to establish the first ever automated MR classification system. Evaluated on a test set of 179 labeled -- 154 non-MR and 25 MR -- sequences, CUSSP attains an F1 score of 0.69 and a ROC-AUC score of 0.88, setting the first benchmark result for this new task.
* 12 pages including references and the appendix. 9 Figures, 2 tables. Accepted at MICCAI (Machine Learning for Automated Mitral Regurgitation Detection from Cardiac Imaging) 2023, Link to Springer at https://link.springer.com/chapter/10.1007/978-3-031-43990-2_23
MultiWave: Multiresolution Deep Architectures through Wavelet Decomposition for Multivariate Time Series Prediction
Jun 16, 2023Abstract:The analysis of multivariate time series data is challenging due to the various frequencies of signal changes that can occur over both short and long terms. Furthermore, standard deep learning models are often unsuitable for such datasets, as signals are typically sampled at different rates. To address these issues, we introduce MultiWave, a novel framework that enhances deep learning time series models by incorporating components that operate at the intrinsic frequencies of signals. MultiWave uses wavelets to decompose each signal into subsignals of varying frequencies and groups them into frequency bands. Each frequency band is handled by a different component of our model. A gating mechanism combines the output of the components to produce sparse models that use only specific signals at specific frequencies. Our experiments demonstrate that MultiWave accurately identifies informative frequency bands and improves the performance of various deep learning models, including LSTM, Transformer, and CNN-based models, for a wide range of applications. It attains top performance in stress and affect detection from wearables. It also increases the AUC of the best-performing model by 5% for in-hospital COVID-19 mortality prediction from patient blood samples and for human activity recognition from accelerometer and gyroscope data. We show that MultiWave consistently identifies critical features and their frequency components, thus providing valuable insights into the applications studied.
Structure Mapping for Transferability of Causal Models
Jul 18, 2020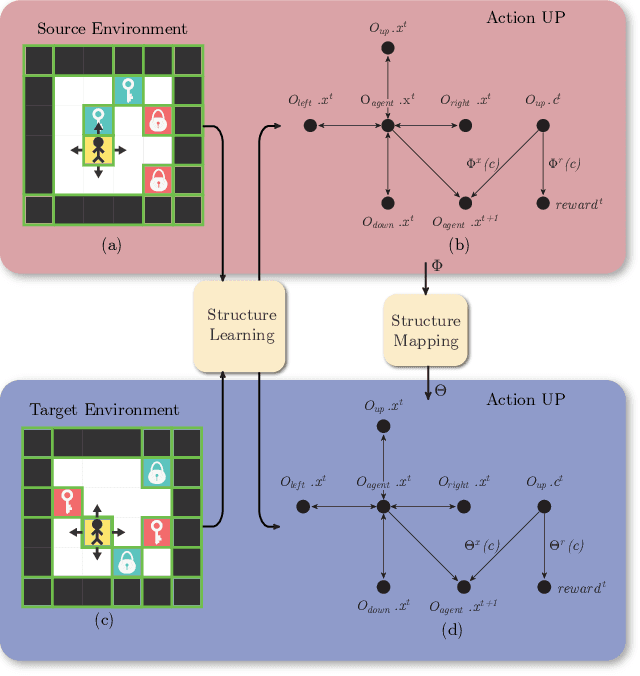
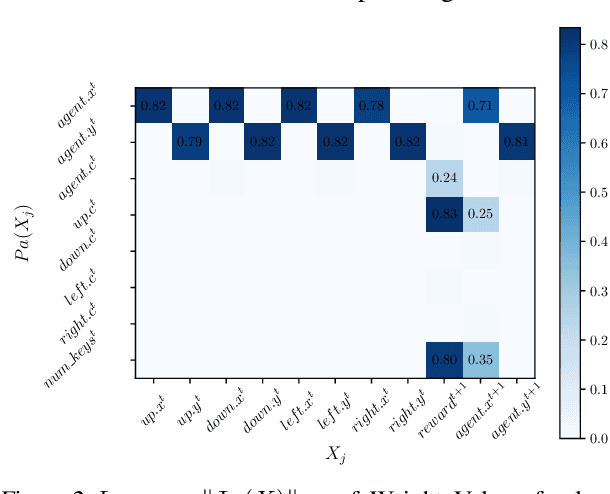
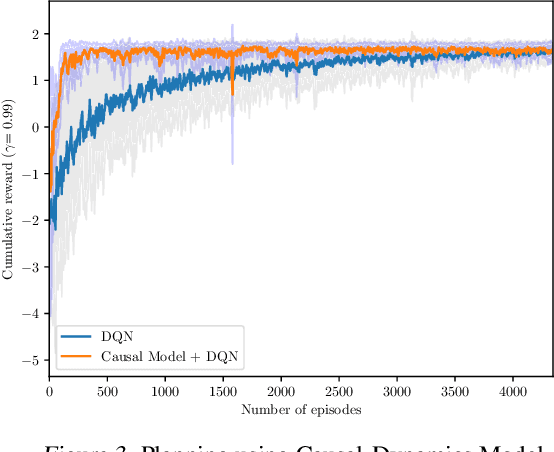
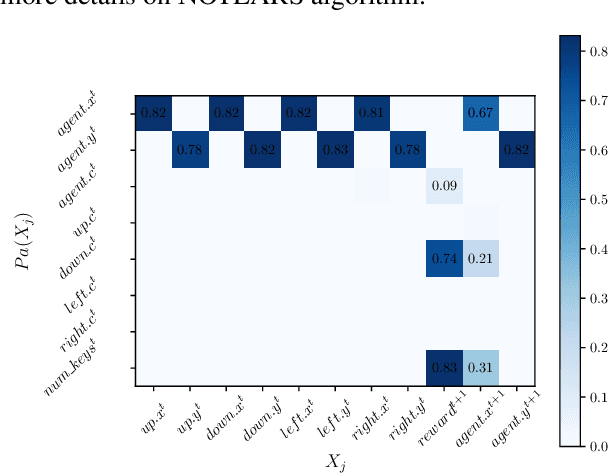
Abstract:Human beings learn causal models and constantly use them to transfer knowledge between similar environments. We use this intuition to design a transfer-learning framework using object-oriented representations to learn the causal relationships between objects. A learned causal dynamics model can be used to transfer between variants of an environment with exchangeable perceptual features among objects but with the same underlying causal dynamics. We adapt continuous optimization for structure learning techniques to explicitly learn the cause and effects of the actions in an interactive environment and transfer to the target domain by categorization of the objects based on causal knowledge. We demonstrate the advantages of our approach in a gridworld setting by combining causal model-based approach with model-free approach in reinforcement learning.
Personalized Student Stress Prediction with Deep Multitask Network
Jun 26, 2019



Abstract:With the growing popularity of wearable devices, the ability to utilize physiological data collected from these devices to predict the wearer's mental state such as mood and stress suggests great clinical applications, yet such a task is extremely challenging. In this paper, we present a general platform for personalized predictive modeling of behavioural states like students' level of stress. Through the use of Auto-encoders and Multitask learning we extend the prediction of stress to both sequences of passive sensor data and high-level covariates. Our model outperforms the state-of-the-art in the prediction of stress level from mobile sensor data, obtaining a 45.6 % improvement in F1 score on the StudentLife dataset.
* 4 Pages, without references. Published in Proceedings of the 1st Adaptive & Multitask Learning Workshop,Long Beach, California, 2019
Alzheimer's Disease Brain MRI Classification: Challenges and Insights
Jun 10, 2019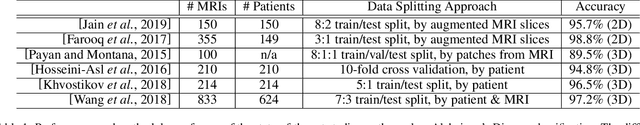
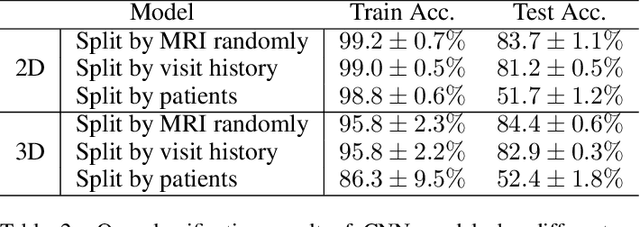
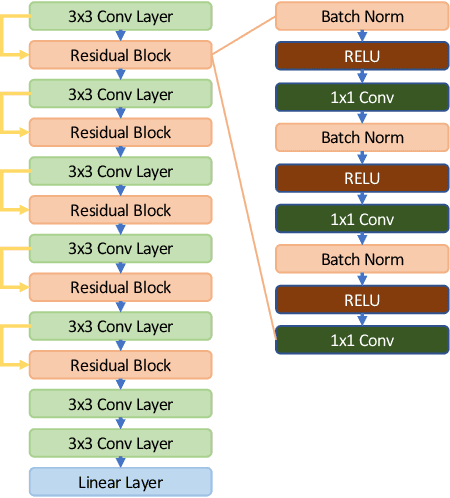
Abstract:In recent years, many papers have reported state-of-the-art performance on Alzheimer's Disease classification with MRI scans from the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset using convolutional neural networks. However, we discover that when we split that data into training and testing sets at the subject level, we are not able to obtain similar performance, bringing the validity of many of the previous studies into question. Furthermore, we point out that previous works use different subsets of the ADNI data, making comparison across similar works tricky. In this study, we present the results of three splitting methods, discuss the motivations behind their validity, and report our results using all of the available subjects.
Multi-resolution Networks For Flexible Irregular Time Series Modeling (Multi-FIT)
Apr 30, 2019



Abstract:Missing values, irregularly collected samples, and multi-resolution signals commonly occur in multivariate time series data, making predictive tasks difficult. These challenges are especially prevalent in the healthcare domain, where patients' vital signs and electronic records are collected at different frequencies and have occasionally missing information due to the imperfections in equipment or patient circumstances. Researchers have handled each of these issues differently, often handling missing data through mean value imputation and then using sequence models over the multivariate signals while ignoring the different resolution of signals. We propose a unified model named Multi-resolution Flexible Irregular Time series Network (Multi-FIT). The building block for Multi-FIT is the FIT network. The FIT network creates an informative dense representation at each time step using signal information such as last observed value, time difference since the last observed time stamp and overall mean for the signal. Vertical FIT (FIT-V) is a variant of FIT which also models the relationship between different temporal signals while creating the informative dense representations for the signal. The multi-FIT model uses multiple FIT networks for sets of signals with different resolutions, further facilitating the construction of flexible representations. Our model has three main contributions: a.) it does not impute values but rather creates informative representations to provide flexibility to the model for creating task-specific representations b.) it models the relationship between different signals in the form of support signals c.) it models different resolutions in parallel before merging them for the final prediction task. The FIT, FIT-V and Multi-FIT networks improve upon the state-of-the-art models for three predictive tasks, including the forecasting of patient survival.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge