Łukasz Kidziński
Department of Biomedical Engineering, Stanford University, California, USA
Open source software for automatic subregional assessment of knee cartilage degradation using quantitative T2 relaxometry and deep learning
Dec 22, 2020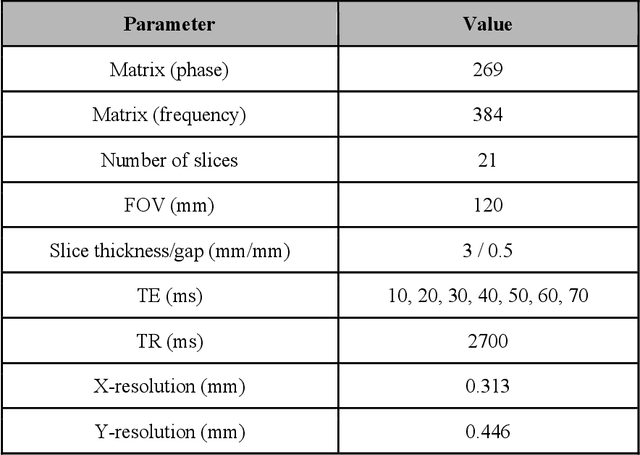
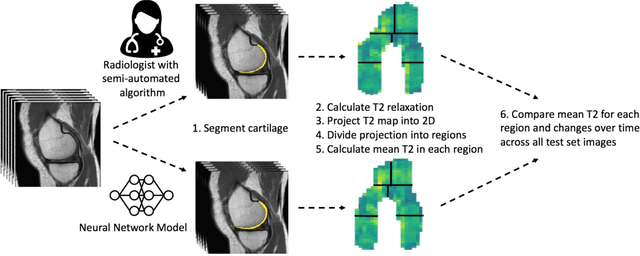
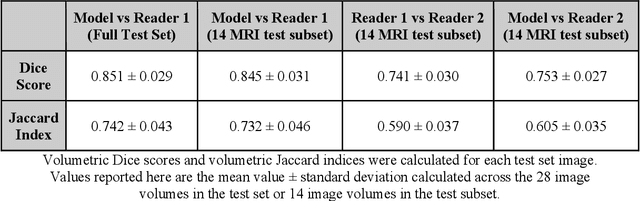
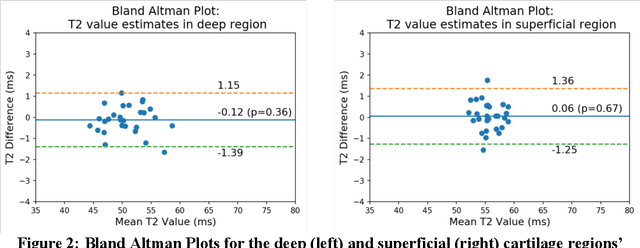
Abstract:Objective: We evaluate a fully-automated femoral cartilage segmentation model for measuring T2 relaxation values and longitudinal changes using multi-echo spin echo (MESE) MRI. We have open sourced this model and corresponding segmentations. Methods: We trained a neural network to segment femoral cartilage from MESE MRIs. Cartilage was divided into 12 subregions along medial-lateral, superficial-deep, and anterior-central-posterior boundaries. Subregional T2 values and four-year changes were calculated using a musculoskeletal radiologist's segmentations (Reader 1) and the model's segmentations. These were compared using 28 held out images. A subset of 14 images were also evaluated by a second expert (Reader 2) for comparison. Results: Model segmentations agreed with Reader 1 segmentations with a Dice score of 0.85 +/- 0.03. The model's estimated T2 values for individual subregions agreed with those of Reader 1 with an average Spearman correlation of 0.89 and average mean absolute error (MAE) of 1.34 ms. The model's estimated four-year change in T2 for individual regions agreed with Reader 1 with an average correlation of 0.80 and average MAE of 1.72 ms. The model agreed with Reader 1 at least as closely as Reader 2 agreed with Reader 1 in terms of Dice score (0.85 vs 0.75) and subregional T2 values. Conclusions: We present a fast, fully-automated model for segmentation of MESE MRIs. Assessments of cartilage health using its segmentations agree with those of an expert as closely as experts agree with one another. This has the potential to accelerate osteoarthritis research.
Generalized Matrix Factorization
Oct 06, 2020



Abstract:Unmeasured or latent variables are often the cause of correlations between multivariate measurements and are studied in a variety of fields such as psychology, ecology, and medicine. For Gaussian measurements, there are classical tools such as factor analysis or principal component analysis with a well-established theory and fast algorithms. Generalized Linear Latent Variable models (GLLVM) generalize such factor models to non-Gaussian responses. However, current algorithms for estimating model parameters in GLLVMs require intensive computation and do not scale to large datasets with thousands of observational units or responses. In this article, we propose a new approach for fitting GLLVMs to such high-volume, high-dimensional datasets. We approximate the likelihood using penalized quasi-likelihood and use a Newton method and Fisher scoring to learn the model parameters. Our method greatly reduces the computation time and can be easily parallelized, enabling factorization at unprecedented scale using commodity hardware. We illustrate application of our method on a dataset of 48,000 observational units with over 2,000 observed species in each unit, finding that most of the variability can be explained with a handful of factors.
Modeling treatment events in disease progression
May 26, 2019



Abstract:Ability to quantify and predict progression of a disease is fundamental for selecting an appropriate treatment. Many clinical metrics cannot be acquired frequently either because of their cost (e.g. MRI, gait analysis) or because they are inconvenient or harmful to a patient (e.g. biopsy, x-ray). In such scenarios, in order to estimate individual trajectories of disease progression, it is advantageous to leverage similarities between patients, i.e. the covariance of trajectories, and find a latent representation of progression. Most of existing methods for estimating trajectories do not account for events in-between observations, what dramatically decreases their adequacy for clinical practice. In this study, we develop a machine learning framework named Coordinatewise-Soft-Impute (CSI) for analyzing disease progression from sparse observations in the presence of confounding events. CSI is guaranteed to converge to the global minimum of the corresponding optimization problem. Experimental results also demonstrates the effectiveness of CSI using both simulated and real dataset.
Artificial Intelligence for Prosthetics - challenge solutions
Feb 07, 2019
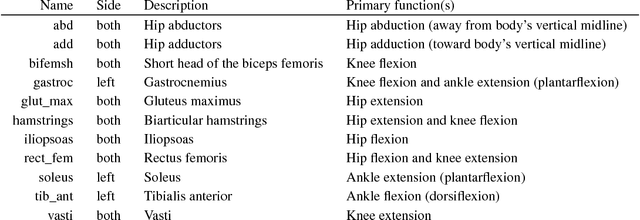
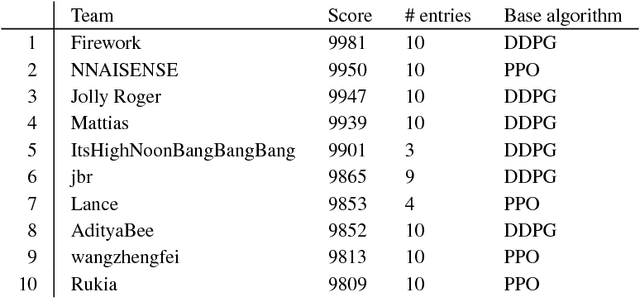
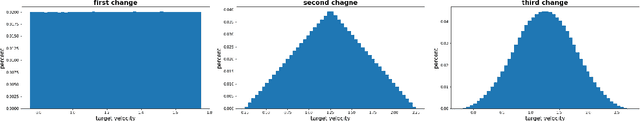
Abstract:In the NeurIPS 2018 Artificial Intelligence for Prosthetics challenge, participants were tasked with building a controller for a musculoskeletal model with a goal of matching a given time-varying velocity vector. Top participants were invited to describe their algorithms. In this work, we describe the challenge and present thirteen solutions that used deep reinforcement learning approaches. Many solutions use similar relaxations and heuristics, such as reward shaping, frame skipping, discretization of the action space, symmetry, and policy blending. However, each team implemented different modifications of the known algorithms by, for example, dividing the task into subtasks, learning low-level control, or by incorporating expert knowledge and using imitation learning.
Longitudinal data analysis using matrix completion
Sep 24, 2018



Abstract:In clinical practice and biomedical research, measurements are often collected sparsely and irregularly in time while the data acquisition is expensive and inconvenient. Examples include measurements of spine bone mineral density, cancer growth through mammography or biopsy, a progression of defect of vision, or assessment of gait in patients with neurological disorders. Since the data collection is often costly and inconvenient, estimation of progression from sparse observations is of great interest for practitioners. From the statistical standpoint, such data is often analyzed in the context of a mixed-effect model where time is treated as both random and fixed effect. Alternatively, researchers analyze Gaussian processes or functional data where observations are assumed to be drawn from a certain distribution of processes. These models are flexible but rely on probabilistic assumptions and require very careful implementation. In this study, we propose an alternative elementary framework for analyzing longitudinal data, relying on matrix completion. Our method yields point estimates of progression curves by iterative application of the SVD. Our framework covers multivariate longitudinal data, regression and can be easily extended to other settings. We apply our methods to understand trends of progression of motor impairment in children with Cerebral Palsy. Our model approximates individual progression curves and explains 30% of the variability. Low-rank representation of progression trends enables discovering that subtypes of Cerebral Palsy exhibit different progression trends.
Learning to Run challenge solutions: Adapting reinforcement learning methods for neuromusculoskeletal environments
Apr 02, 2018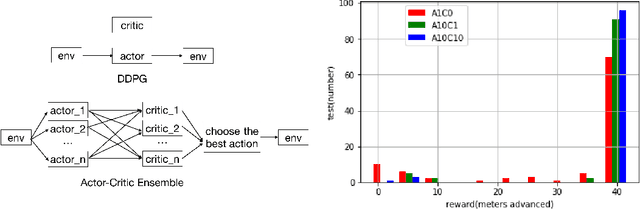
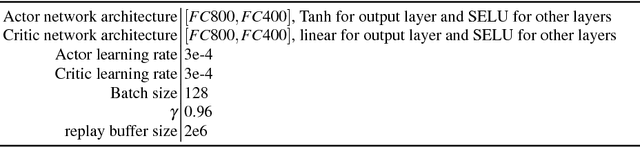
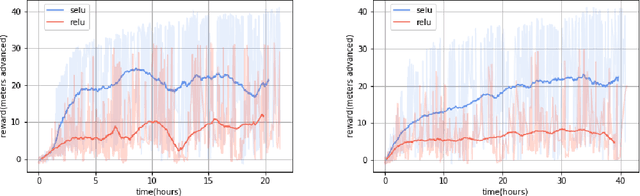
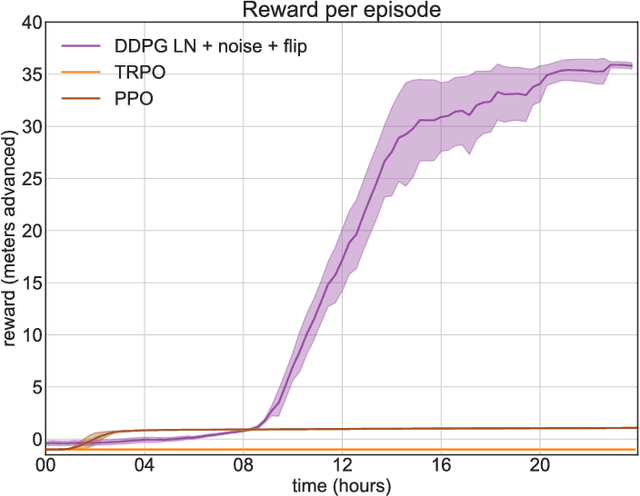
Abstract:In the NIPS 2017 Learning to Run challenge, participants were tasked with building a controller for a musculoskeletal model to make it run as fast as possible through an obstacle course. Top participants were invited to describe their algorithms. In this work, we present eight solutions that used deep reinforcement learning approaches, based on algorithms such as Deep Deterministic Policy Gradient, Proximal Policy Optimization, and Trust Region Policy Optimization. Many solutions use similar relaxations and heuristics, such as reward shaping, frame skipping, discretization of the action space, symmetry, and policy blending. However, each of the eight teams implemented different modifications of the known algorithms.
Learning to Run challenge: Synthesizing physiologically accurate motion using deep reinforcement learning
Mar 31, 2018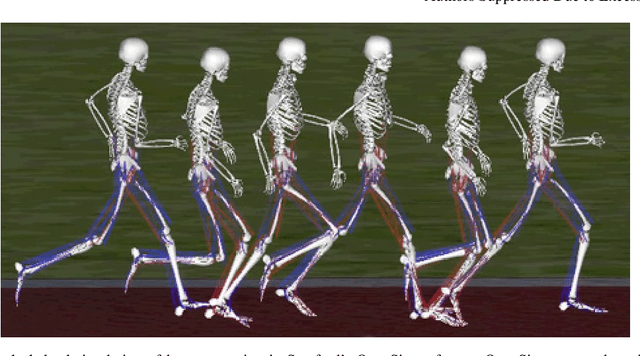
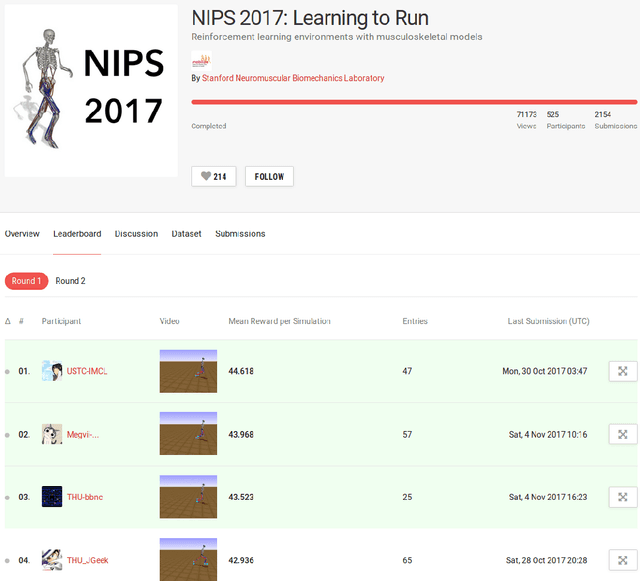
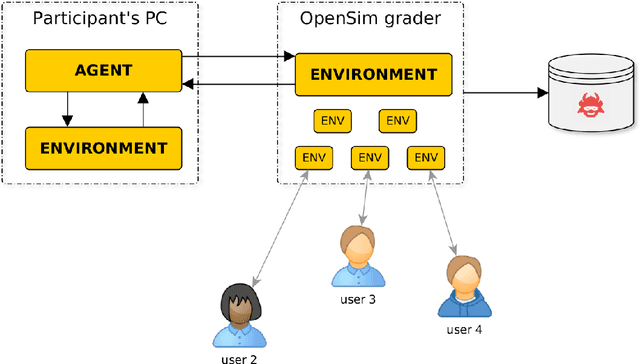
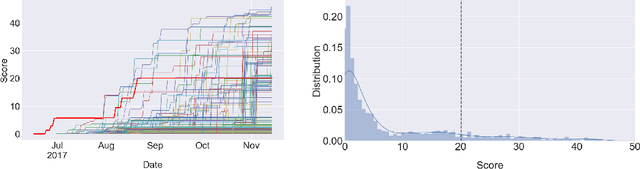
Abstract:Synthesizing physiologically-accurate human movement in a variety of conditions can help practitioners plan surgeries, design experiments, or prototype assistive devices in simulated environments, reducing time and costs and improving treatment outcomes. Because of the large and complex solution spaces of biomechanical models, current methods are constrained to specific movements and models, requiring careful design of a controller and hindering many possible applications. We sought to discover if modern optimization methods efficiently explore these complex spaces. To do this, we posed the problem as a competition in which participants were tasked with developing a controller to enable a physiologically-based human model to navigate a complex obstacle course as quickly as possible, without using any experimental data. They were provided with a human musculoskeletal model and a physics-based simulation environment. In this paper, we discuss the design of the competition, technical difficulties, results, and analysis of the top controllers. The challenge proved that deep reinforcement learning techniques, despite their high computational cost, can be successfully employed as an optimization method for synthesizing physiologically feasible motion in high-dimensional biomechanical systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge