Ron Keuth
Fracture Morphology Classification: Local Multiclass Modeling for Multilabel Complexity
Dec 16, 2025

Abstract:Between $15\,\%$ and $45\,\%$ of children experience a fracture during their growth years, making accurate diagnosis essential. Fracture morphology, alongside location and fragment angle, is a key diagnostic feature. In this work, we propose a method to extract fracture morphology by assigning automatically global AO codes to corresponding fracture bounding boxes. This approach enables the use of public datasets and reformulates the global multilabel task into a local multiclass one, improving the average F1 score by $7.89\,\%$. However, performance declines when using imperfect fracture detectors, highlighting challenges for real-world deployment. Our code is available on GitHub.
BronchoGAN: Anatomically consistent and domain-agnostic image-to-image translation for video bronchoscopy
Jul 02, 2025Abstract:The limited availability of bronchoscopy images makes image synthesis particularly interesting for training deep learning models. Robust image translation across different domains -- virtual bronchoscopy, phantom as well as in-vivo and ex-vivo image data -- is pivotal for clinical applications. This paper proposes BronchoGAN introducing anatomical constraints for image-to-image translation being integrated into a conditional GAN. In particular, we force bronchial orifices to match across input and output images. We further propose to use foundation model-generated depth images as intermediate representation ensuring robustness across a variety of input domains establishing models with substantially less reliance on individual training datasets. Moreover our intermediate depth image representation allows to easily construct paired image data for training. Our experiments showed that input images from different domains (e.g. virtual bronchoscopy, phantoms) can be successfully translated to images mimicking realistic human airway appearance. We demonstrated that anatomical settings (i.e. bronchial orifices) can be robustly preserved with our approach which is shown qualitatively and quantitatively by means of improved FID, SSIM and dice coefficients scores. Our anatomical constraints enabled an improvement in the Dice coefficient of up to 0.43 for synthetic images. Through foundation models for intermediate depth representations, bronchial orifice segmentation integrated as anatomical constraints into conditional GANs we are able to robustly translate images from different bronchoscopy input domains. BronchoGAN allows to incorporate public CT scan data (virtual bronchoscopy) in order to generate large-scale bronchoscopy image datasets with realistic appearance. BronchoGAN enables to bridge the gap of missing public bronchoscopy images.
A Systematic Analysis of Input Modalities for Fracture Classification of the Paediatric Wrist
Dec 18, 2024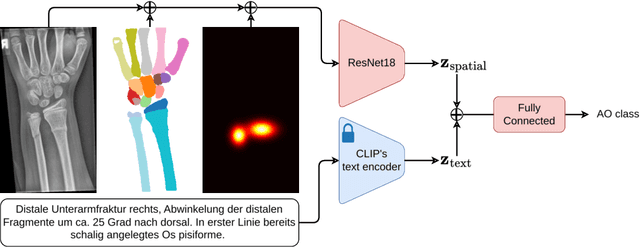
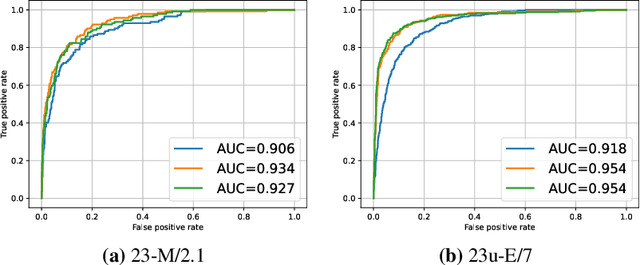
Abstract:Fractures, particularly in the distal forearm, are among the most common injuries in children and adolescents, with approximately 800 000 cases treated annually in Germany. The AO/OTA system provides a structured fracture type classification, which serves as the foundation for treatment decisions. Although accurately classifying fractures can be challenging, current deep learning models have demonstrated performance comparable to that of experienced radiologists. While most existing approaches rely solely on radiographs, the potential impact of incorporating other additional modalities, such as automatic bone segmentation, fracture location, and radiology reports, remains underexplored. In this work, we systematically analyse the contribution of these three additional information types, finding that combining them with radiographs increases the AUROC from 91.71 to 93.25. Our code is available on GitHub.
SAM Carries the Burden: A Semi-Supervised Approach Refining Pseudo Labels for Medical Segmentation
Nov 19, 2024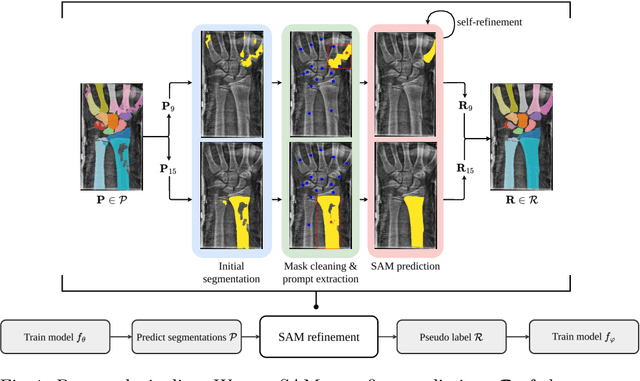
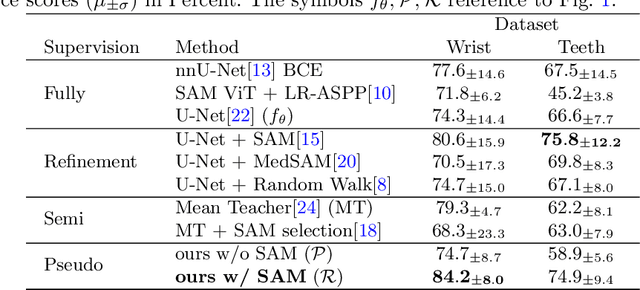
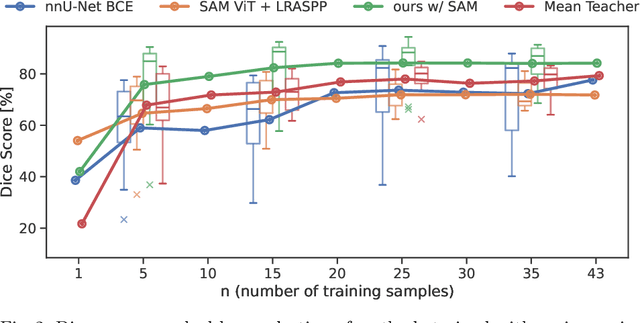
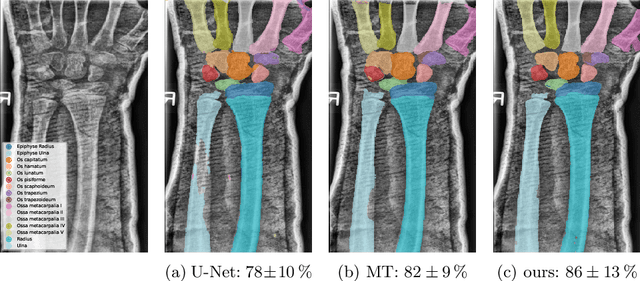
Abstract:Semantic segmentation is a crucial task in medical imaging. Although supervised learning techniques have proven to be effective in performing this task, they heavily depend on large amounts of annotated training data. The recently introduced Segment Anything Model (SAM) enables prompt-based segmentation and offers zero-shot generalization to unfamiliar objects. In our work, we leverage SAM's abstract object understanding for medical image segmentation to provide pseudo labels for semi-supervised learning, thereby mitigating the need for extensive annotated training data. Our approach refines initial segmentations that are derived from a limited amount of annotated data (comprising up to 43 cases) by extracting bounding boxes and seed points as prompts forwarded to SAM. Thus, it enables the generation of dense segmentation masks as pseudo labels for unlabelled data. The results show that training with our pseudo labels yields an improvement in Dice score from $74.29\,\%$ to $84.17\,\%$ and from $66.63\,\%$ to $74.87\,\%$ for the segmentation of bones of the paediatric wrist and teeth in dental radiographs, respectively. As a result, our method outperforms intensity-based post-processing methods, state-of-the-art supervised learning for segmentation (nnU-Net), and the semi-supervised mean teacher approach. Our Code is available on GitHub.
DenseSeg: Joint Learning for Semantic Segmentation and Landmark Detection Using Dense Image-to-Shape Representation
May 30, 2024



Abstract:Purpose: Semantic segmentation and landmark detection are fundamental tasks of medical image processing, facilitating further analysis of anatomical objects. Although deep learning-based pixel-wise classification has set a new-state-of-the-art for segmentation, it falls short in landmark detection, a strength of shape-based approaches. Methods: In this work, we propose a dense image-to-shape representation that enables the joint learning of landmarks and semantic segmentation by employing a fully convolutional architecture. Our method intuitively allows the extraction of arbitrary landmarks due to its representation of anatomical correspondences. We benchmark our method against the state-of-the-art for semantic segmentation (nnUNet), a shape-based approach employing geometric deep learning and a CNN-based method for landmark detection. Results: We evaluate our method on two medical dataset: one common benchmark featuring the lungs, heart, and clavicle from thorax X-rays, and another with 17 different bones in the paediatric wrist. While our method is on pair with the landmark detection baseline in the thorax setting (error in mm of $2.6\pm0.9$ vs $2.7\pm0.9$), it substantially surpassed it in the more complex wrist setting ($1.1\pm0.6$ vs $1.9\pm0.5$). Conclusion: We demonstrate that dense geometric shape representation is beneficial for challenging landmark detection tasks and outperforms previous state-of-the-art using heatmap regression. While it does not require explicit training on the landmarks themselves, allowing for the addition of new landmarks without necessitating retraining.}
Combining Image- and Geometric-based Deep Learning for Shape Regression: A Comparison to Pixel-level Methods for Segmentation in Chest X-Ray
Jan 15, 2024Abstract:When solving a segmentation task, shaped-base methods can be beneficial compared to pixelwise classification due to geometric understanding of the target object as shape, preventing the generation of anatomical implausible predictions in particular for corrupted data. In this work, we propose a novel hybrid method that combines a lightweight CNN backbone with a geometric neural network (Point Transformer) for shape regression. Using the same CNN encoder, the Point Transformer reaches segmentation quality on per with current state-of-the-art convolutional decoders ($4\pm1.9$ vs $3.9\pm2.9$ error in mm and $85\pm13$ vs $88\pm10$ Dice), but crucially, is more stable w.r.t image distortion, starting to outperform them at a corruption level of 30%. Furthermore, we include the nnU-Net as an upper baseline, which has $3.7\times$ more trainable parameters than our proposed method.
Airway Label Prediction in Video Bronchoscopy: Capturing Temporal Dependencies Utilizing Anatomical Knowledge
Jul 17, 2023Abstract:Purpose: Navigation guidance is a key requirement for a multitude of lung interventions using video bronchoscopy. State-of-the-art solutions focus on lung biopsies using electromagnetic tracking and intraoperative image registration w.r.t. preoperative CT scans for guidance. The requirement of patient-specific CT scans hampers the utilisation of navigation guidance for other applications such as intensive care units. Methods: This paper addresses navigation guidance solely incorporating bronchosopy video data. In contrast to state-of-the-art approaches we entirely omit the use of electromagnetic tracking and patient-specific CT scans. Guidance is enabled by means of topological bronchoscope localization w.r.t. an interpatient airway model. Particularly, we take maximally advantage of anatomical constraints of airway trees being sequentially traversed. This is realized by incorporating sequences of CNN-based airway likelihoods into a Hidden Markov Model. Results: Our approach is evaluated based on multiple experiments inside a lung phantom model. With the consideration of temporal context and use of anatomical knowledge for regularization, we are able to improve the accuracy up to to 0.98 compared to 0.81 (weighted F1: 0.98 compared to 0.81) for a classification based on individual frames. Conclusion: We combine CNN-based single image classification of airway segments with anatomical constraints and temporal HMM-based inference for the first time. Our approach renders vision-only guidance for bronchoscopy interventions in the absence of electromagnetic tracking and patient-specific CT scans possible.
Weakly Supervised Airway Orifice Segmentation in Video Bronchoscopy
Aug 24, 2022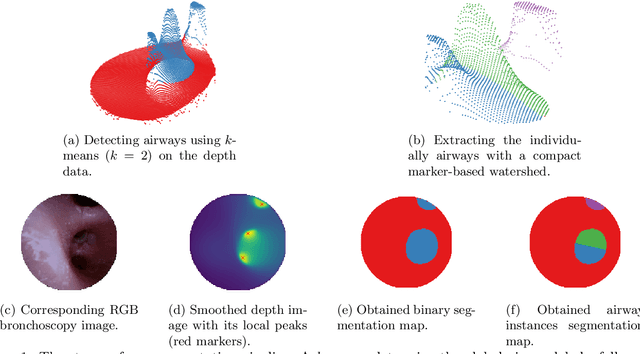
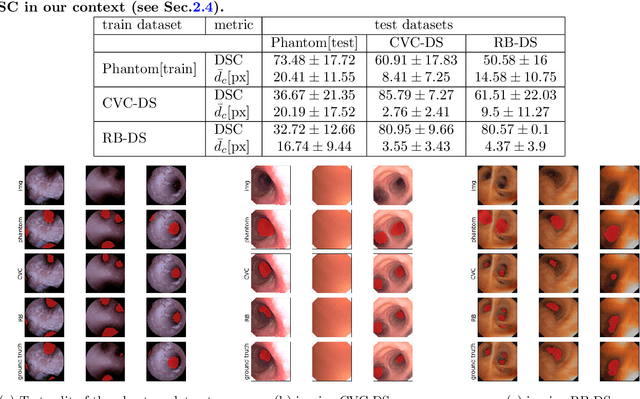
Abstract:Video bronchoscopy is routinely conducted for biopsies of lung tissue suspected for cancer, monitoring of COPD patients and clarification of acute respiratory problems at intensive care units. The navigation within complex bronchial trees is particularly challenging and physically demanding, requiring long-term experiences of physicians. This paper addresses the automatic segmentation of bronchial orifices in bronchoscopy videos. Deep learning-based approaches to this task are currently hampered due to the lack of readily-available ground truth segmentation data. Thus, we present a data-driven pipeline consisting of a k-means followed by a compact marker-based watershed algorithm which enables to generate airway instance segmentation maps from given depth images. In this way, these traditional algorithms serve as weak supervision for training a shallow CNN directly on RGB images solely based on a phantom dataset. We evaluate generalization capabilities of this model on two in-vivo datasets covering 250 frames on 21 different bronchoscopies. We demonstrate that its performance is comparable to those models being directly trained on in-vivo data, reaching an average error of 11 vs 5 pixels for the detected centers of the airway segmentation by an image resolution of 128x128. Our quantitative and qualitative results indicate that in the context of video bronchoscopy, phantom data and weak supervision using non-learning-based approaches enable to gain a semantic understanding of airway structures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge