Ludger Tüshaus
A Systematic Analysis of Input Modalities for Fracture Classification of the Paediatric Wrist
Dec 18, 2024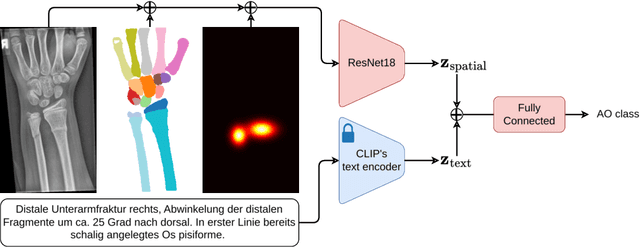
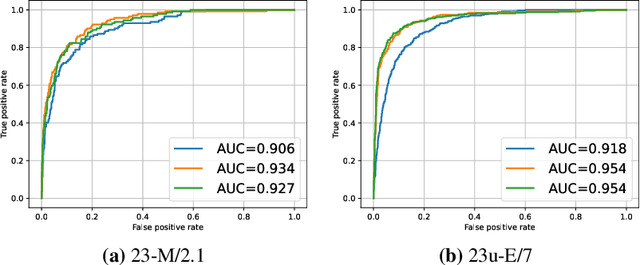
Abstract:Fractures, particularly in the distal forearm, are among the most common injuries in children and adolescents, with approximately 800 000 cases treated annually in Germany. The AO/OTA system provides a structured fracture type classification, which serves as the foundation for treatment decisions. Although accurately classifying fractures can be challenging, current deep learning models have demonstrated performance comparable to that of experienced radiologists. While most existing approaches rely solely on radiographs, the potential impact of incorporating other additional modalities, such as automatic bone segmentation, fracture location, and radiology reports, remains underexplored. In this work, we systematically analyse the contribution of these three additional information types, finding that combining them with radiographs increases the AUROC from 91.71 to 93.25. Our code is available on GitHub.
SAM Carries the Burden: A Semi-Supervised Approach Refining Pseudo Labels for Medical Segmentation
Nov 19, 2024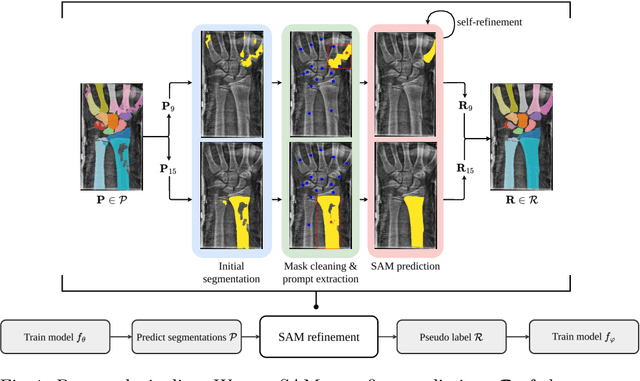
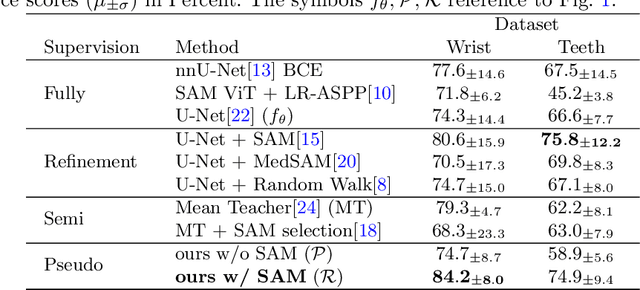
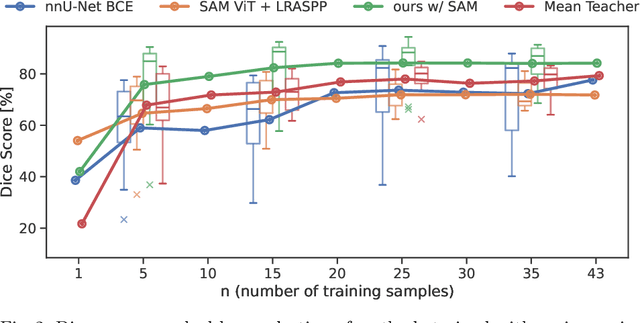
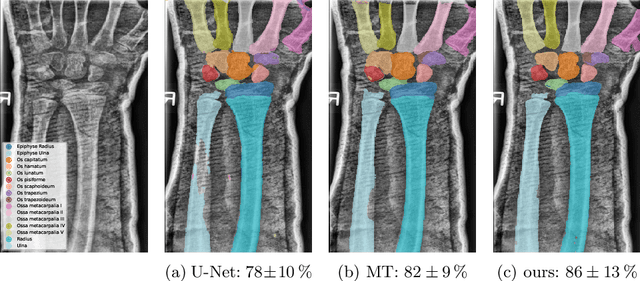
Abstract:Semantic segmentation is a crucial task in medical imaging. Although supervised learning techniques have proven to be effective in performing this task, they heavily depend on large amounts of annotated training data. The recently introduced Segment Anything Model (SAM) enables prompt-based segmentation and offers zero-shot generalization to unfamiliar objects. In our work, we leverage SAM's abstract object understanding for medical image segmentation to provide pseudo labels for semi-supervised learning, thereby mitigating the need for extensive annotated training data. Our approach refines initial segmentations that are derived from a limited amount of annotated data (comprising up to 43 cases) by extracting bounding boxes and seed points as prompts forwarded to SAM. Thus, it enables the generation of dense segmentation masks as pseudo labels for unlabelled data. The results show that training with our pseudo labels yields an improvement in Dice score from $74.29\,\%$ to $84.17\,\%$ and from $66.63\,\%$ to $74.87\,\%$ for the segmentation of bones of the paediatric wrist and teeth in dental radiographs, respectively. As a result, our method outperforms intensity-based post-processing methods, state-of-the-art supervised learning for segmentation (nnU-Net), and the semi-supervised mean teacher approach. Our Code is available on GitHub.
DenseSeg: Joint Learning for Semantic Segmentation and Landmark Detection Using Dense Image-to-Shape Representation
May 30, 2024



Abstract:Purpose: Semantic segmentation and landmark detection are fundamental tasks of medical image processing, facilitating further analysis of anatomical objects. Although deep learning-based pixel-wise classification has set a new-state-of-the-art for segmentation, it falls short in landmark detection, a strength of shape-based approaches. Methods: In this work, we propose a dense image-to-shape representation that enables the joint learning of landmarks and semantic segmentation by employing a fully convolutional architecture. Our method intuitively allows the extraction of arbitrary landmarks due to its representation of anatomical correspondences. We benchmark our method against the state-of-the-art for semantic segmentation (nnUNet), a shape-based approach employing geometric deep learning and a CNN-based method for landmark detection. Results: We evaluate our method on two medical dataset: one common benchmark featuring the lungs, heart, and clavicle from thorax X-rays, and another with 17 different bones in the paediatric wrist. While our method is on pair with the landmark detection baseline in the thorax setting (error in mm of $2.6\pm0.9$ vs $2.7\pm0.9$), it substantially surpassed it in the more complex wrist setting ($1.1\pm0.6$ vs $1.9\pm0.5$). Conclusion: We demonstrate that dense geometric shape representation is beneficial for challenging landmark detection tasks and outperforms previous state-of-the-art using heatmap regression. While it does not require explicit training on the landmarks themselves, allowing for the addition of new landmarks without necessitating retraining.}
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge