Mattias Heinrich
Non-Invasive 3D Wound Measurement with RGB-D Imaging
Jan 26, 2026Abstract:Chronic wound monitoring and management require accurate and efficient wound measurement methods. This paper presents a fast, non-invasive 3D wound measurement algorithm based on RGB-D imaging. The method combines RGB-D odometry with B-spline surface reconstruction to generate detailed 3D wound meshes, enabling automatic computation of clinically relevant wound measurements such as perimeter, surface area, and dimensions. We evaluated our system on realistic silicone wound phantoms and measured sub-millimetre 3D reconstruction accuracy compared with high-resolution ground-truth scans. The extracted measurements demonstrated low variability across repeated captures and strong agreement with manual assessments. The proposed pipeline also outperformed a state-of-the-art object-centric RGB-D reconstruction method while maintaining runtimes suitable for real-time clinical deployment. Our approach offers a promising tool for automated wound assessment in both clinical and remote healthcare settings.
A Systematic Analysis of Input Modalities for Fracture Classification of the Paediatric Wrist
Dec 18, 2024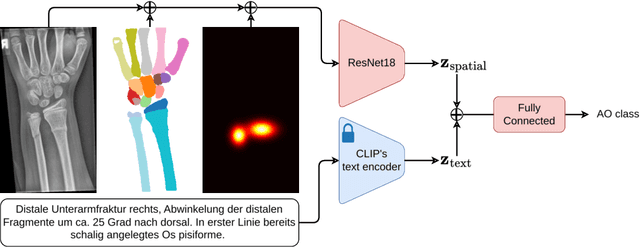
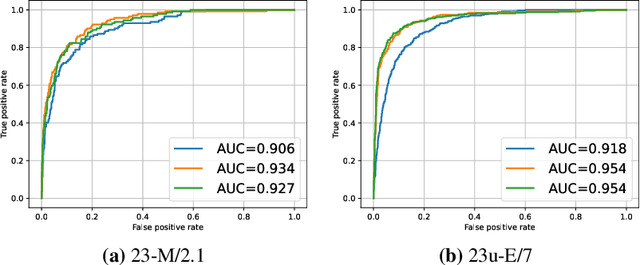
Abstract:Fractures, particularly in the distal forearm, are among the most common injuries in children and adolescents, with approximately 800 000 cases treated annually in Germany. The AO/OTA system provides a structured fracture type classification, which serves as the foundation for treatment decisions. Although accurately classifying fractures can be challenging, current deep learning models have demonstrated performance comparable to that of experienced radiologists. While most existing approaches rely solely on radiographs, the potential impact of incorporating other additional modalities, such as automatic bone segmentation, fracture location, and radiology reports, remains underexplored. In this work, we systematically analyse the contribution of these three additional information types, finding that combining them with radiographs increases the AUROC from 91.71 to 93.25. Our code is available on GitHub.
SAM Carries the Burden: A Semi-Supervised Approach Refining Pseudo Labels for Medical Segmentation
Nov 19, 2024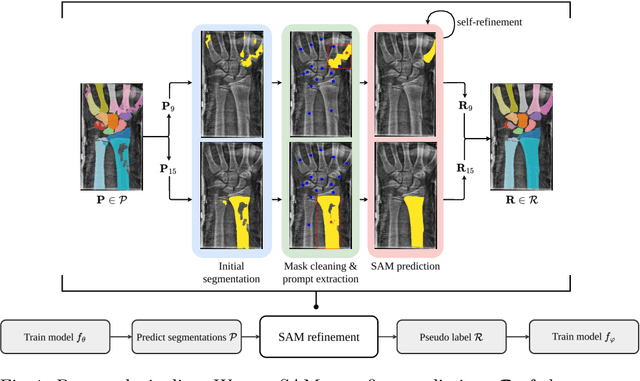
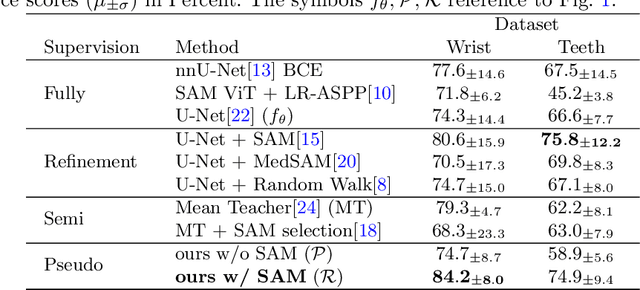
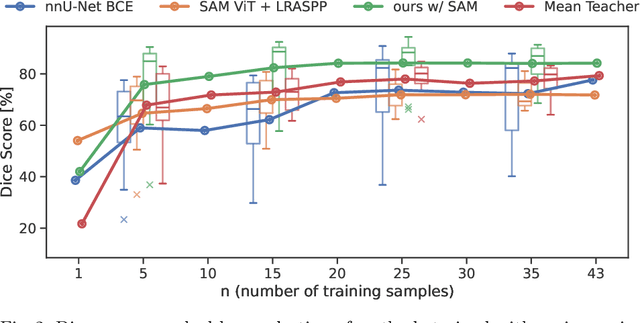
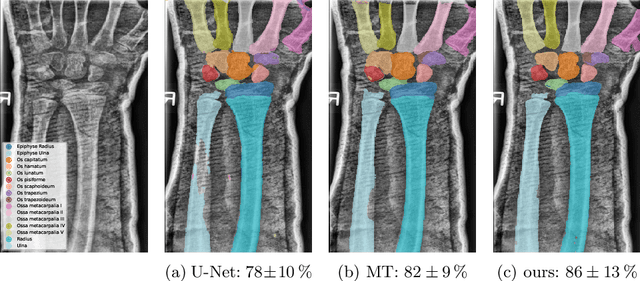
Abstract:Semantic segmentation is a crucial task in medical imaging. Although supervised learning techniques have proven to be effective in performing this task, they heavily depend on large amounts of annotated training data. The recently introduced Segment Anything Model (SAM) enables prompt-based segmentation and offers zero-shot generalization to unfamiliar objects. In our work, we leverage SAM's abstract object understanding for medical image segmentation to provide pseudo labels for semi-supervised learning, thereby mitigating the need for extensive annotated training data. Our approach refines initial segmentations that are derived from a limited amount of annotated data (comprising up to 43 cases) by extracting bounding boxes and seed points as prompts forwarded to SAM. Thus, it enables the generation of dense segmentation masks as pseudo labels for unlabelled data. The results show that training with our pseudo labels yields an improvement in Dice score from $74.29\,\%$ to $84.17\,\%$ and from $66.63\,\%$ to $74.87\,\%$ for the segmentation of bones of the paediatric wrist and teeth in dental radiographs, respectively. As a result, our method outperforms intensity-based post-processing methods, state-of-the-art supervised learning for segmentation (nnU-Net), and the semi-supervised mean teacher approach. Our Code is available on GitHub.
DenseSeg: Joint Learning for Semantic Segmentation and Landmark Detection Using Dense Image-to-Shape Representation
May 30, 2024



Abstract:Purpose: Semantic segmentation and landmark detection are fundamental tasks of medical image processing, facilitating further analysis of anatomical objects. Although deep learning-based pixel-wise classification has set a new-state-of-the-art for segmentation, it falls short in landmark detection, a strength of shape-based approaches. Methods: In this work, we propose a dense image-to-shape representation that enables the joint learning of landmarks and semantic segmentation by employing a fully convolutional architecture. Our method intuitively allows the extraction of arbitrary landmarks due to its representation of anatomical correspondences. We benchmark our method against the state-of-the-art for semantic segmentation (nnUNet), a shape-based approach employing geometric deep learning and a CNN-based method for landmark detection. Results: We evaluate our method on two medical dataset: one common benchmark featuring the lungs, heart, and clavicle from thorax X-rays, and another with 17 different bones in the paediatric wrist. While our method is on pair with the landmark detection baseline in the thorax setting (error in mm of $2.6\pm0.9$ vs $2.7\pm0.9$), it substantially surpassed it in the more complex wrist setting ($1.1\pm0.6$ vs $1.9\pm0.5$). Conclusion: We demonstrate that dense geometric shape representation is beneficial for challenging landmark detection tasks and outperforms previous state-of-the-art using heatmap regression. While it does not require explicit training on the landmarks themselves, allowing for the addition of new landmarks without necessitating retraining.}
Combining Image- and Geometric-based Deep Learning for Shape Regression: A Comparison to Pixel-level Methods for Segmentation in Chest X-Ray
Jan 15, 2024Abstract:When solving a segmentation task, shaped-base methods can be beneficial compared to pixelwise classification due to geometric understanding of the target object as shape, preventing the generation of anatomical implausible predictions in particular for corrupted data. In this work, we propose a novel hybrid method that combines a lightweight CNN backbone with a geometric neural network (Point Transformer) for shape regression. Using the same CNN encoder, the Point Transformer reaches segmentation quality on per with current state-of-the-art convolutional decoders ($4\pm1.9$ vs $3.9\pm2.9$ error in mm and $85\pm13$ vs $88\pm10$ Dice), but crucially, is more stable w.r.t image distortion, starting to outperform them at a corruption level of 30%. Furthermore, we include the nnU-Net as an upper baseline, which has $3.7\times$ more trainable parameters than our proposed method.
Airway Label Prediction in Video Bronchoscopy: Capturing Temporal Dependencies Utilizing Anatomical Knowledge
Jul 17, 2023Abstract:Purpose: Navigation guidance is a key requirement for a multitude of lung interventions using video bronchoscopy. State-of-the-art solutions focus on lung biopsies using electromagnetic tracking and intraoperative image registration w.r.t. preoperative CT scans for guidance. The requirement of patient-specific CT scans hampers the utilisation of navigation guidance for other applications such as intensive care units. Methods: This paper addresses navigation guidance solely incorporating bronchosopy video data. In contrast to state-of-the-art approaches we entirely omit the use of electromagnetic tracking and patient-specific CT scans. Guidance is enabled by means of topological bronchoscope localization w.r.t. an interpatient airway model. Particularly, we take maximally advantage of anatomical constraints of airway trees being sequentially traversed. This is realized by incorporating sequences of CNN-based airway likelihoods into a Hidden Markov Model. Results: Our approach is evaluated based on multiple experiments inside a lung phantom model. With the consideration of temporal context and use of anatomical knowledge for regularization, we are able to improve the accuracy up to to 0.98 compared to 0.81 (weighted F1: 0.98 compared to 0.81) for a classification based on individual frames. Conclusion: We combine CNN-based single image classification of airway segments with anatomical constraints and temporal HMM-based inference for the first time. Our approach renders vision-only guidance for bronchoscopy interventions in the absence of electromagnetic tracking and patient-specific CT scans possible.
Why is the winner the best?
Mar 30, 2023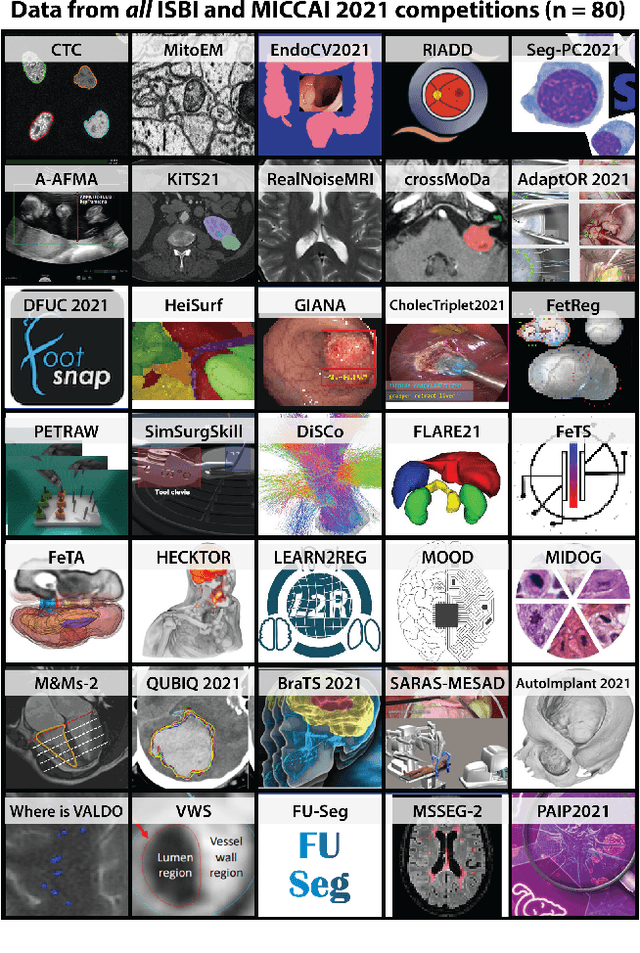
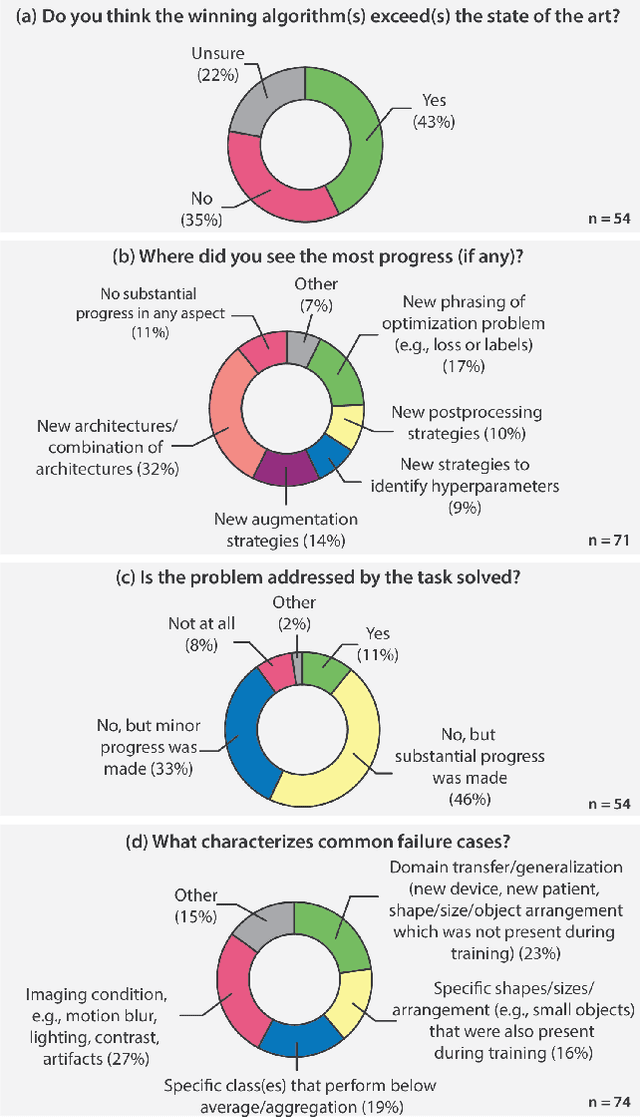
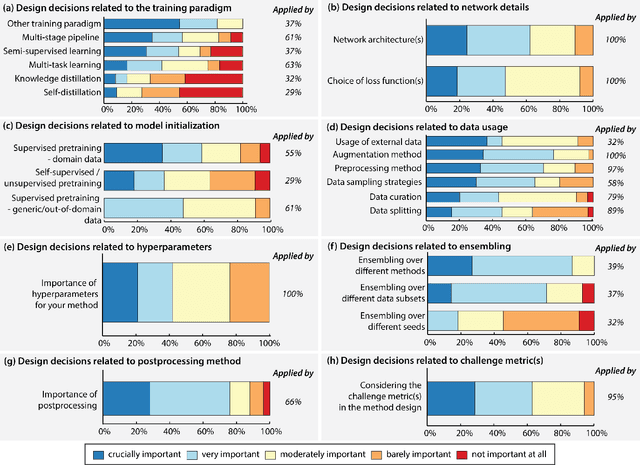
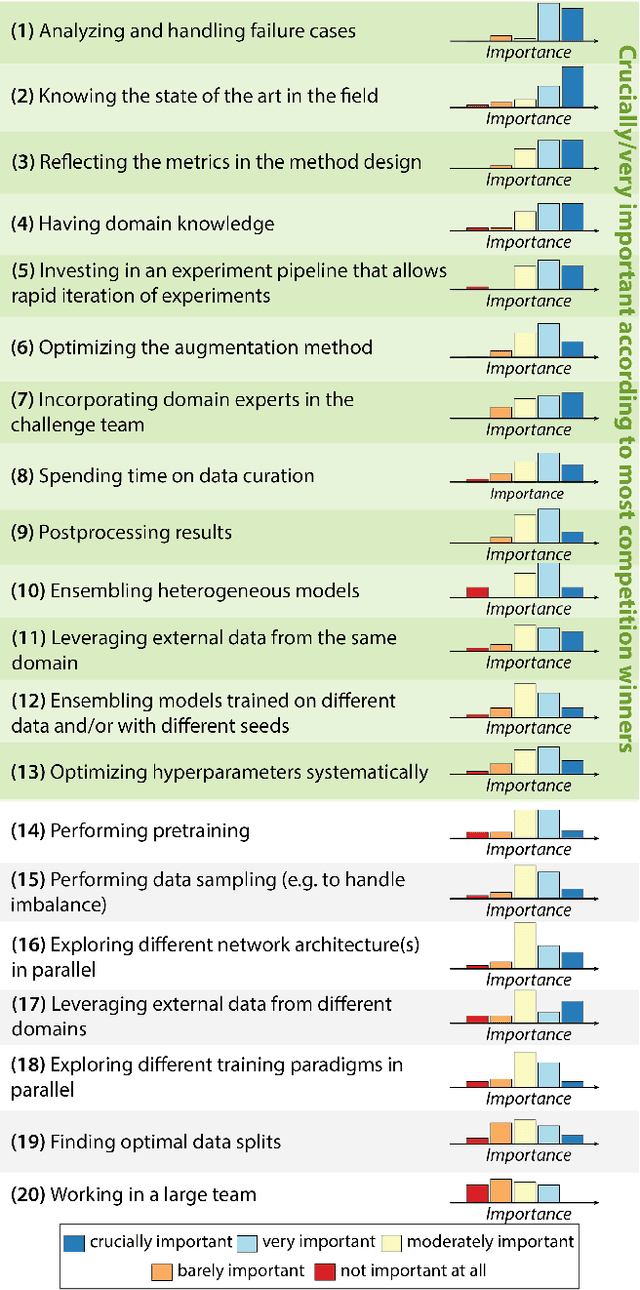
Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Weakly Supervised Airway Orifice Segmentation in Video Bronchoscopy
Aug 24, 2022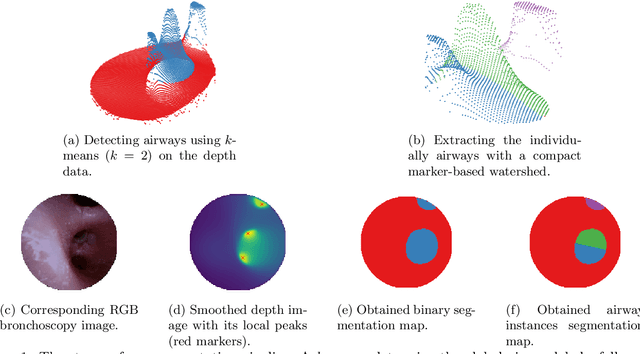
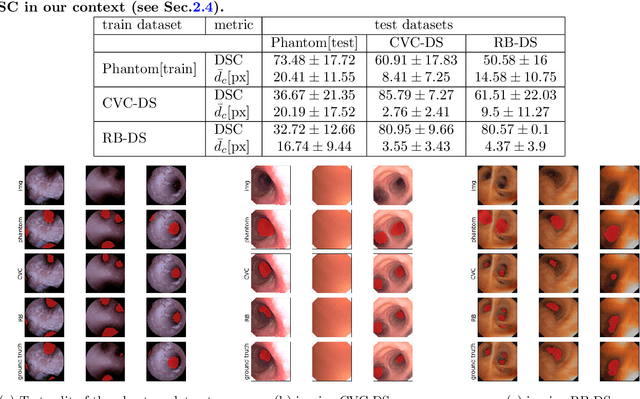
Abstract:Video bronchoscopy is routinely conducted for biopsies of lung tissue suspected for cancer, monitoring of COPD patients and clarification of acute respiratory problems at intensive care units. The navigation within complex bronchial trees is particularly challenging and physically demanding, requiring long-term experiences of physicians. This paper addresses the automatic segmentation of bronchial orifices in bronchoscopy videos. Deep learning-based approaches to this task are currently hampered due to the lack of readily-available ground truth segmentation data. Thus, we present a data-driven pipeline consisting of a k-means followed by a compact marker-based watershed algorithm which enables to generate airway instance segmentation maps from given depth images. In this way, these traditional algorithms serve as weak supervision for training a shallow CNN directly on RGB images solely based on a phantom dataset. We evaluate generalization capabilities of this model on two in-vivo datasets covering 250 frames on 21 different bronchoscopies. We demonstrate that its performance is comparable to those models being directly trained on in-vivo data, reaching an average error of 11 vs 5 pixels for the detected centers of the airway segmentation by an image resolution of 128x128. Our quantitative and qualitative results indicate that in the context of video bronchoscopy, phantom data and weak supervision using non-learning-based approaches enable to gain a semantic understanding of airway structures.
Learn2Trust: A video and streamlit-based educational programme for AI-based medical image analysis targeted towards medical students
Aug 15, 2022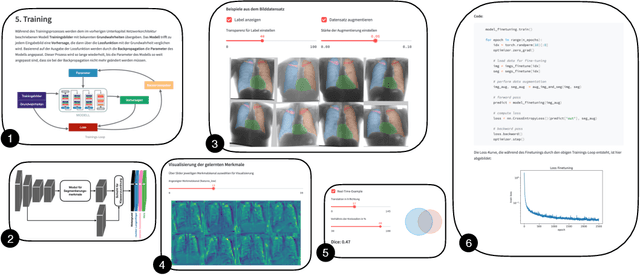
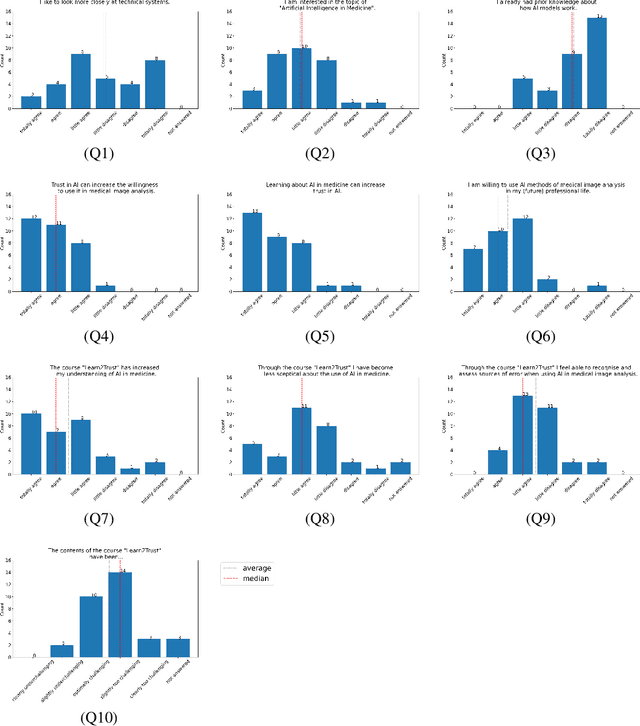
Abstract:In order to be able to use artificial intelligence (AI) in medicine without scepticism and to recognise and assess its growing potential, a basic understanding of this topic is necessary among current and future medical staff. Under the premise of "trust through understanding", we developed an innovative online course as a learning opportunity within the framework of the German KI Campus (AI campus) project, which is a self-guided course that teaches the basics of AI for the analysis of medical image data. The main goal is to provide a learning environment for a sufficient understanding of AI in medical image analysis so that further interest in this topic is stimulated and inhibitions towards its use can be overcome by means of positive application experience. The focus was on medical applications and the fundamentals of machine learning. The online course was divided into consecutive lessons, which include theory in the form of explanatory videos, practical exercises in the form of Streamlit and practical exercises and/or quizzes to check learning progress. A survey among the participating medical students in the first run of the course was used to analyse our research hypotheses quantitatively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge