Marian Himstedt
BronchoGAN: Anatomically consistent and domain-agnostic image-to-image translation for video bronchoscopy
Jul 02, 2025Abstract:The limited availability of bronchoscopy images makes image synthesis particularly interesting for training deep learning models. Robust image translation across different domains -- virtual bronchoscopy, phantom as well as in-vivo and ex-vivo image data -- is pivotal for clinical applications. This paper proposes BronchoGAN introducing anatomical constraints for image-to-image translation being integrated into a conditional GAN. In particular, we force bronchial orifices to match across input and output images. We further propose to use foundation model-generated depth images as intermediate representation ensuring robustness across a variety of input domains establishing models with substantially less reliance on individual training datasets. Moreover our intermediate depth image representation allows to easily construct paired image data for training. Our experiments showed that input images from different domains (e.g. virtual bronchoscopy, phantoms) can be successfully translated to images mimicking realistic human airway appearance. We demonstrated that anatomical settings (i.e. bronchial orifices) can be robustly preserved with our approach which is shown qualitatively and quantitatively by means of improved FID, SSIM and dice coefficients scores. Our anatomical constraints enabled an improvement in the Dice coefficient of up to 0.43 for synthetic images. Through foundation models for intermediate depth representations, bronchial orifice segmentation integrated as anatomical constraints into conditional GANs we are able to robustly translate images from different bronchoscopy input domains. BronchoGAN allows to incorporate public CT scan data (virtual bronchoscopy) in order to generate large-scale bronchoscopy image datasets with realistic appearance. BronchoGAN enables to bridge the gap of missing public bronchoscopy images.
Classification of Prostate Cancer in 3D Magnetic Resonance Imaging Data based on Convolutional Neural Networks
Apr 16, 2024Abstract:Prostate cancer is a commonly diagnosed cancerous disease among men world-wide. Even with modern technology such as multi-parametric magnetic resonance tomography and guided biopsies, the process for diagnosing prostate cancer remains time consuming and requires highly trained professionals. In this paper, different convolutional neural networks (CNN) are evaluated on their abilities to reliably classify whether an MRI sequence contains malignant lesions. Implementations of a ResNet, a ConvNet and a ConvNeXt for 3D image data are trained and evaluated. The models are trained using different data augmentation techniques, learning rates, and optimizers. The data is taken from a private dataset, provided by Cantonal Hospital Aarau. The best result was achieved by a ResNet3D, yielding an average precision score of 0.4583 and AUC ROC score of 0.6214.
Airway Label Prediction in Video Bronchoscopy: Capturing Temporal Dependencies Utilizing Anatomical Knowledge
Jul 17, 2023Abstract:Purpose: Navigation guidance is a key requirement for a multitude of lung interventions using video bronchoscopy. State-of-the-art solutions focus on lung biopsies using electromagnetic tracking and intraoperative image registration w.r.t. preoperative CT scans for guidance. The requirement of patient-specific CT scans hampers the utilisation of navigation guidance for other applications such as intensive care units. Methods: This paper addresses navigation guidance solely incorporating bronchosopy video data. In contrast to state-of-the-art approaches we entirely omit the use of electromagnetic tracking and patient-specific CT scans. Guidance is enabled by means of topological bronchoscope localization w.r.t. an interpatient airway model. Particularly, we take maximally advantage of anatomical constraints of airway trees being sequentially traversed. This is realized by incorporating sequences of CNN-based airway likelihoods into a Hidden Markov Model. Results: Our approach is evaluated based on multiple experiments inside a lung phantom model. With the consideration of temporal context and use of anatomical knowledge for regularization, we are able to improve the accuracy up to to 0.98 compared to 0.81 (weighted F1: 0.98 compared to 0.81) for a classification based on individual frames. Conclusion: We combine CNN-based single image classification of airway segments with anatomical constraints and temporal HMM-based inference for the first time. Our approach renders vision-only guidance for bronchoscopy interventions in the absence of electromagnetic tracking and patient-specific CT scans possible.
Weakly Supervised Airway Orifice Segmentation in Video Bronchoscopy
Aug 24, 2022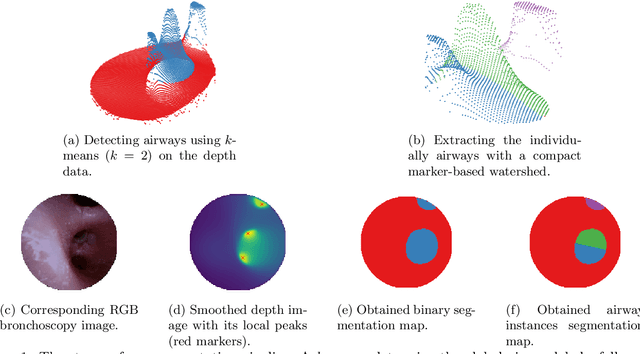
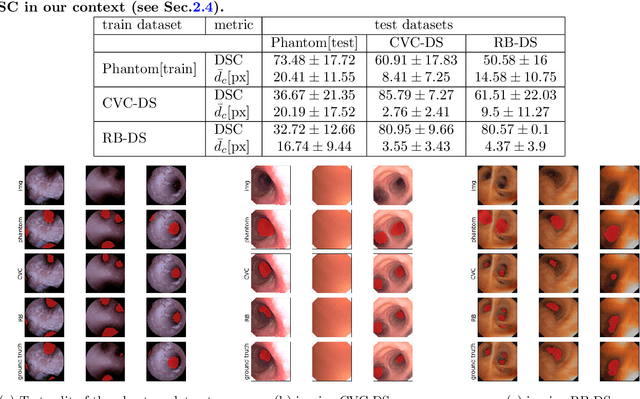
Abstract:Video bronchoscopy is routinely conducted for biopsies of lung tissue suspected for cancer, monitoring of COPD patients and clarification of acute respiratory problems at intensive care units. The navigation within complex bronchial trees is particularly challenging and physically demanding, requiring long-term experiences of physicians. This paper addresses the automatic segmentation of bronchial orifices in bronchoscopy videos. Deep learning-based approaches to this task are currently hampered due to the lack of readily-available ground truth segmentation data. Thus, we present a data-driven pipeline consisting of a k-means followed by a compact marker-based watershed algorithm which enables to generate airway instance segmentation maps from given depth images. In this way, these traditional algorithms serve as weak supervision for training a shallow CNN directly on RGB images solely based on a phantom dataset. We evaluate generalization capabilities of this model on two in-vivo datasets covering 250 frames on 21 different bronchoscopies. We demonstrate that its performance is comparable to those models being directly trained on in-vivo data, reaching an average error of 11 vs 5 pixels for the detected centers of the airway segmentation by an image resolution of 128x128. Our quantitative and qualitative results indicate that in the context of video bronchoscopy, phantom data and weak supervision using non-learning-based approaches enable to gain a semantic understanding of airway structures.
Learn2Trust: A video and streamlit-based educational programme for AI-based medical image analysis targeted towards medical students
Aug 15, 2022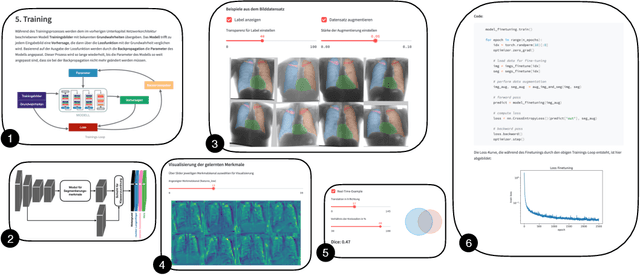
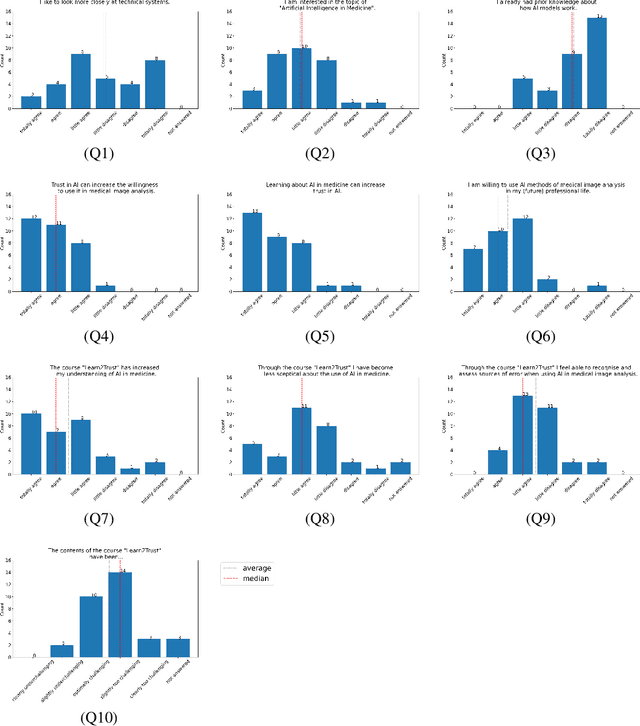
Abstract:In order to be able to use artificial intelligence (AI) in medicine without scepticism and to recognise and assess its growing potential, a basic understanding of this topic is necessary among current and future medical staff. Under the premise of "trust through understanding", we developed an innovative online course as a learning opportunity within the framework of the German KI Campus (AI campus) project, which is a self-guided course that teaches the basics of AI for the analysis of medical image data. The main goal is to provide a learning environment for a sufficient understanding of AI in medical image analysis so that further interest in this topic is stimulated and inhibitions towards its use can be overcome by means of positive application experience. The focus was on medical applications and the fundamentals of machine learning. The online course was divided into consecutive lessons, which include theory in the form of explanatory videos, practical exercises in the form of Streamlit and practical exercises and/or quizzes to check learning progress. A survey among the participating medical students in the first run of the course was used to analyse our research hypotheses quantitatively.
Automatic Generation of Synthetic Colonoscopy Videos for Domain Randomization
May 20, 2022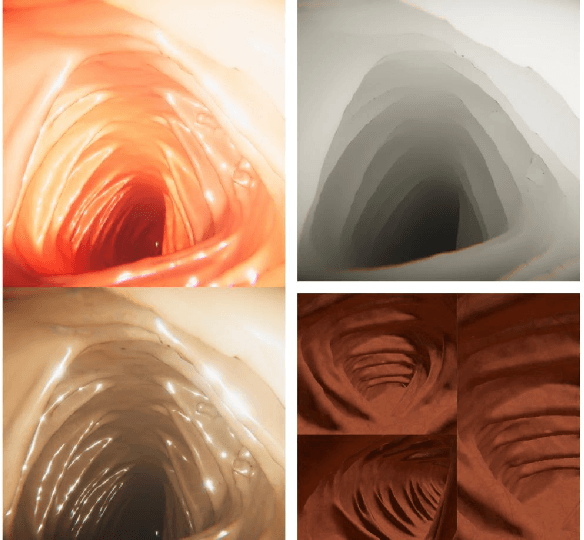
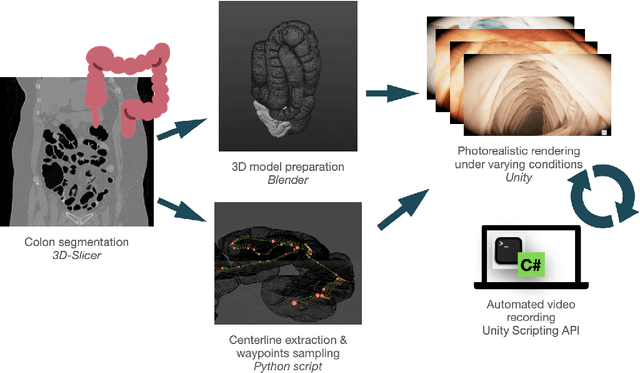
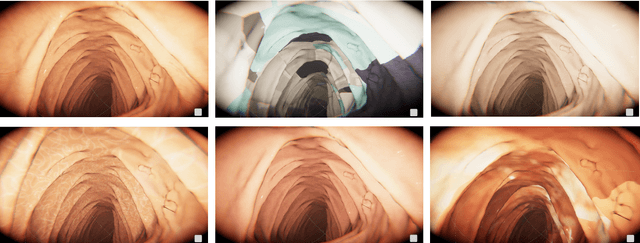
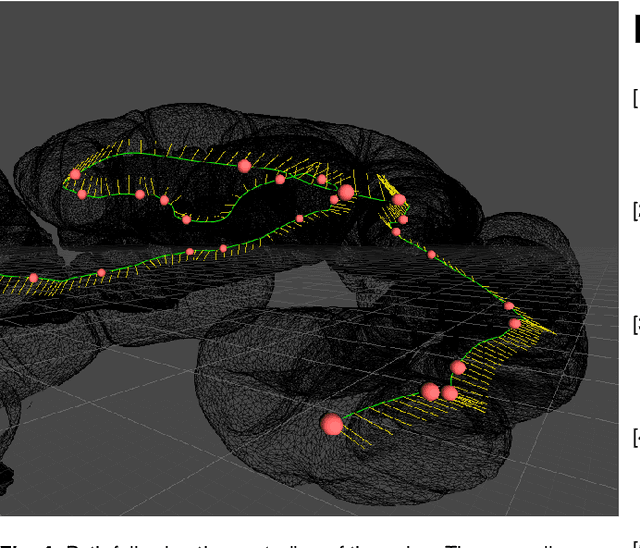
Abstract:An increasing number of colonoscopic guidance and assistance systems rely on machine learning algorithms which require a large amount of high-quality training data. In order to ensure high performance, the latter has to resemble a substantial portion of possible configurations. This particularly addresses varying anatomy, mucosa appearance and image sensor characteristics which are likely deteriorated by motion blur and inadequate illumination. The limited amount of readily available training data hampers to account for all of these possible configurations which results in reduced generalization capabilities of machine learning models. We propose an exemplary solution for synthesizing colonoscopy videos with substantial appearance and anatomical variations which enables to learn discriminative domain-randomized representations of the interior colon while mimicking real-world settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge