Gorkem Can Ates
Med3DVLM: An Efficient Vision-Language Model for 3D Medical Image Analysis
Mar 25, 2025Abstract:Vision-language models (VLMs) have shown promise in 2D medical image analysis, but extending them to 3D remains challenging due to the high computational demands of volumetric data and the difficulty of aligning 3D spatial features with clinical text. We present Med3DVLM, a 3D VLM designed to address these challenges through three key innovations: (1) DCFormer, an efficient encoder that uses decomposed 3D convolutions to capture fine-grained spatial features at scale; (2) SigLIP, a contrastive learning strategy with pairwise sigmoid loss that improves image-text alignment without relying on large negative batches; and (3) a dual-stream MLP-Mixer projector that fuses low- and high-level image features with text embeddings for richer multi-modal representations. We evaluate our model on the M3D dataset, which includes radiology reports and VQA data for 120,084 3D medical images. Results show that Med3DVLM achieves superior performance across multiple benchmarks. For image-text retrieval, it reaches 61.00% R@1 on 2,000 samples, significantly outperforming the current state-of-the-art M3D model (19.10%). For report generation, it achieves a METEOR score of 36.42% (vs. 14.38%). In open-ended visual question answering (VQA), it scores 36.76% METEOR (vs. 33.58%), and in closed-ended VQA, it achieves 79.95% accuracy (vs. 75.78%). These results highlight Med3DVLM's ability to bridge the gap between 3D imaging and language, enabling scalable, multi-task reasoning across clinical applications. Our code is publicly available at https://github.com/mirthAI/Med3DVLM.
DCFormer: Efficient 3D Vision-Language Modeling with Decomposed Convolutions
Feb 07, 2025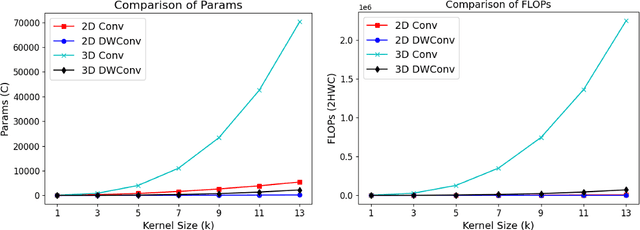



Abstract:Vision-language models (VLMs) align visual and textual representations, enabling high-performance zero-shot classification and image-text retrieval in 2D medical imaging. However, extending VLMs to 3D medical imaging remains computationally challenging. Existing 3D VLMs rely on Vision Transformers (ViTs), which are computationally expensive due to self-attention's quadratic complexity, or 3D convolutions, which demand excessive parameters and FLOPs as kernel size increases. We introduce DCFormer, an efficient 3D medical image encoder that factorizes 3D convolutions into three parallel 1D convolutions along depth, height, and width. This design preserves spatial information while significantly reducing computational cost. Integrated into a CLIP-based vision-language framework, DCFormer is evaluated on CT-RATE, a dataset of 50,188 paired 3D chest CT volumes and radiology reports, for zero-shot multi-abnormality detection across 18 pathologies. Compared to ViT, ConvNeXt, PoolFormer, and TransUNet, DCFormer achieves superior efficiency and accuracy, with DCFormer-Tiny reaching 62.0% accuracy and a 46.3% F1-score while using significantly fewer parameters. These results highlight DCFormer's potential for scalable, clinically deployable 3D medical VLMs. Our codes will be publicly available.
Dual Cross-Attention for Medical Image Segmentation
Mar 30, 2023



Abstract:We propose Dual Cross-Attention (DCA), a simple yet effective attention module that is able to enhance skip-connections in U-Net-based architectures for medical image segmentation. DCA addresses the semantic gap between encoder and decoder features by sequentially capturing channel and spatial dependencies across multi-scale encoder features. First, the Channel Cross-Attention (CCA) extracts global channel-wise dependencies by utilizing cross-attention across channel tokens of multi-scale encoder features. Then, the Spatial Cross-Attention (SCA) module performs cross-attention to capture spatial dependencies across spatial tokens. Finally, these fine-grained encoder features are up-sampled and connected to their corresponding decoder parts to form the skip-connection scheme. Our proposed DCA module can be integrated into any encoder-decoder architecture with skip-connections such as U-Net and its variants. We test our DCA module by integrating it into six U-Net-based architectures such as U-Net, V-Net, R2Unet, ResUnet++, DoubleUnet and MultiResUnet. Our DCA module shows Dice Score improvements up to 2.05% on GlaS, 2.74% on MoNuSeg, 1.37% on CVC-ClinicDB, 1.12% on Kvasir-Seg and 1.44% on Synapse datasets. Our codes are available at: https://github.com/gorkemcanates/Dual-Cross-Attention
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge