Richard Fan
K2-Think: A Parameter-Efficient Reasoning System
Sep 09, 2025Abstract:K2-Think is a reasoning system that achieves state-of-the-art performance with a 32B parameter model, matching or surpassing much larger models like GPT-OSS 120B and DeepSeek v3.1. Built on the Qwen2.5 base model, our system shows that smaller models can compete at the highest levels by combining advanced post-training and test-time computation techniques. The approach is based on six key technical pillars: Long Chain-of-thought Supervised Finetuning, Reinforcement Learning with Verifiable Rewards (RLVR), Agentic planning prior to reasoning, Test-time Scaling, Speculative Decoding, and Inference-optimized Hardware, all using publicly available open-source datasets. K2-Think excels in mathematical reasoning, achieving state-of-the-art scores on public benchmarks for open-source models, while also performing strongly in other areas such as Code and Science. Our results confirm that a more parameter-efficient model like K2-Think 32B can compete with state-of-the-art systems through an integrated post-training recipe that includes long chain-of-thought training and strategic inference-time enhancements, making open-source reasoning systems more accessible and affordable. K2-Think is freely available at k2think.ai, offering best-in-class inference speeds of over 2,000 tokens per second per request via the Cerebras Wafer-Scale Engine.
Revisiting Reinforcement Learning for LLM Reasoning from A Cross-Domain Perspective
Jun 17, 2025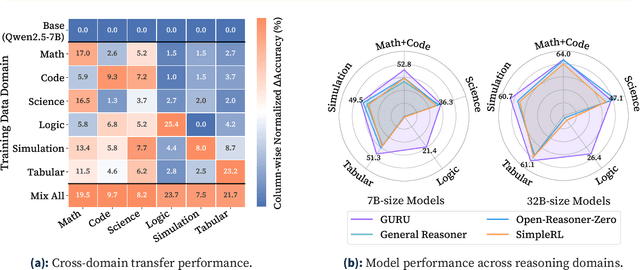
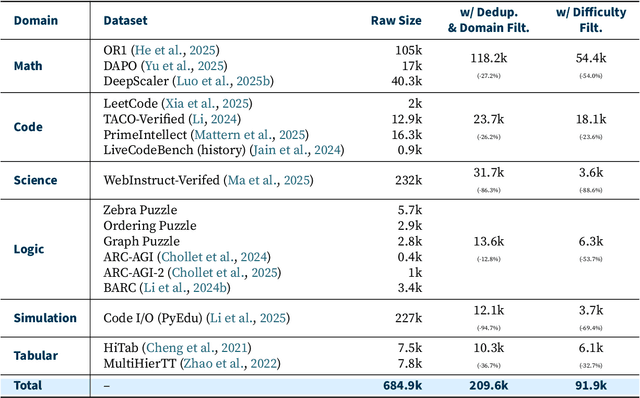
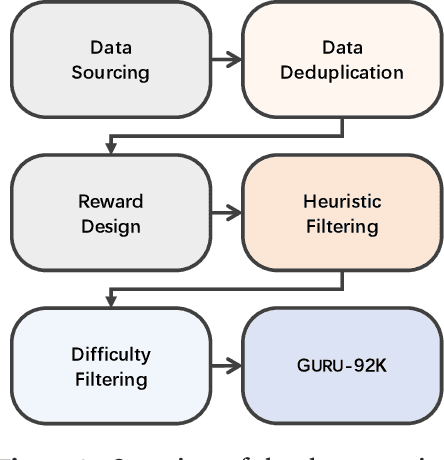
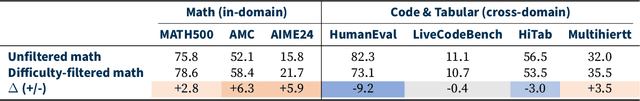
Abstract:Reinforcement learning (RL) has emerged as a promising approach to improve large language model (LLM) reasoning, yet most open efforts focus narrowly on math and code, limiting our understanding of its broader applicability to general reasoning. A key challenge lies in the lack of reliable, scalable RL reward signals across diverse reasoning domains. We introduce Guru, a curated RL reasoning corpus of 92K verifiable examples spanning six reasoning domains--Math, Code, Science, Logic, Simulation, and Tabular--each built through domain-specific reward design, deduplication, and filtering to ensure reliability and effectiveness for RL training. Based on Guru, we systematically revisit established findings in RL for LLM reasoning and observe significant variation across domains. For example, while prior work suggests that RL primarily elicits existing knowledge from pretrained models, our results reveal a more nuanced pattern: domains frequently seen during pretraining (Math, Code, Science) easily benefit from cross-domain RL training, while domains with limited pretraining exposure (Logic, Simulation, and Tabular) require in-domain training to achieve meaningful performance gains, suggesting that RL is likely to facilitate genuine skill acquisition. Finally, we present Guru-7B and Guru-32B, two models that achieve state-of-the-art performance among open models RL-trained with publicly available data, outperforming best baselines by 7.9% and 6.7% on our 17-task evaluation suite across six reasoning domains. We also show that our models effectively improve the Pass@k performance of their base models, particularly on complex tasks less likely to appear in pretraining data. We release data, models, training and evaluation code to facilitate general-purpose reasoning at: https://github.com/LLM360/Reasoning360
Registration-Enhanced Segmentation Method for Prostate Cancer in Ultrasound Images
Feb 02, 2025



Abstract:Prostate cancer is a major cause of cancer-related deaths in men, where early detection greatly improves survival rates. Although MRI-TRUS fusion biopsy offers superior accuracy by combining MRI's detailed visualization with TRUS's real-time guidance, it is a complex and time-intensive procedure that relies heavily on manual annotations, leading to potential errors. To address these challenges, we propose a fully automatic MRI-TRUS fusion-based segmentation method that identifies prostate tumors directly in TRUS images without requiring manual annotations. Unlike traditional multimodal fusion approaches that rely on naive data concatenation, our method integrates a registration-segmentation framework to align and leverage spatial information between MRI and TRUS modalities. This alignment enhances segmentation accuracy and reduces reliance on manual effort. Our approach was validated on a dataset of 1,747 patients from Stanford Hospital, achieving an average Dice coefficient of 0.212, outperforming TRUS-only (0.117) and naive MRI-TRUS fusion (0.132) methods, with significant improvements (p $<$ 0.01). This framework demonstrates the potential for reducing the complexity of prostate cancer diagnosis and provides a flexible architecture applicable to other multimodal medical imaging tasks.
LLM360 K2: Building a 65B 360-Open-Source Large Language Model from Scratch
Jan 16, 2025



Abstract:We detail the training of the LLM360 K2-65B model, scaling up our 360-degree OPEN SOURCE approach to the largest and most powerful models under project LLM360. While open-source LLMs continue to advance, the answer to "How are the largest LLMs trained?" remains unclear within the community. The implementation details for such high-capacity models are often protected due to business considerations associated with their high cost. This lack of transparency prevents LLM researchers from leveraging valuable insights from prior experience, e.g., "What are the best practices for addressing loss spikes?" The LLM360 K2 project addresses this gap by providing full transparency and access to resources accumulated during the training of LLMs at the largest scale. This report highlights key elements of the K2 project, including our first model, K2 DIAMOND, a 65 billion-parameter LLM that surpasses LLaMA-65B and rivals LLaMA2-70B, while requiring fewer FLOPs and tokens. We detail the implementation steps and present a longitudinal analysis of K2 DIAMOND's capabilities throughout its training process. We also outline ongoing projects such as TXT360, setting the stage for future models in the series. By offering previously unavailable resources, the K2 project also resonates with the 360-degree OPEN SOURCE principles of transparency, reproducibility, and accessibility, which we believe are vital in the era of resource-intensive AI research.
LLM360: Towards Fully Transparent Open-Source LLMs
Dec 11, 2023



Abstract:The recent surge in open-source Large Language Models (LLMs), such as LLaMA, Falcon, and Mistral, provides diverse options for AI practitioners and researchers. However, most LLMs have only released partial artifacts, such as the final model weights or inference code, and technical reports increasingly limit their scope to high-level design choices and surface statistics. These choices hinder progress in the field by degrading transparency into the training of LLMs and forcing teams to rediscover many details in the training process. We present LLM360, an initiative to fully open-source LLMs, which advocates for all training code and data, model checkpoints, and intermediate results to be made available to the community. The goal of LLM360 is to support open and collaborative AI research by making the end-to-end LLM training process transparent and reproducible by everyone. As a first step of LLM360, we release two 7B parameter LLMs pre-trained from scratch, Amber and CrystalCoder, including their training code, data, intermediate checkpoints, and analyses (at https://www.llm360.ai). We are committed to continually pushing the boundaries of LLMs through this open-source effort. More large-scale and stronger models are underway and will be released in the future.
Critical Evaluation of Artificial Intelligence as Digital Twin of Pathologist for Prostate Cancer Pathology
Aug 23, 2023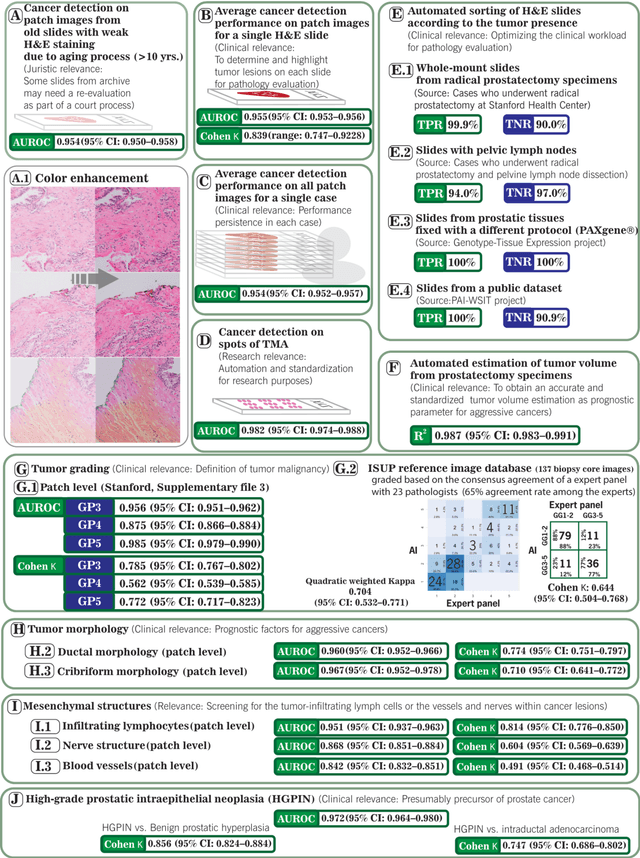
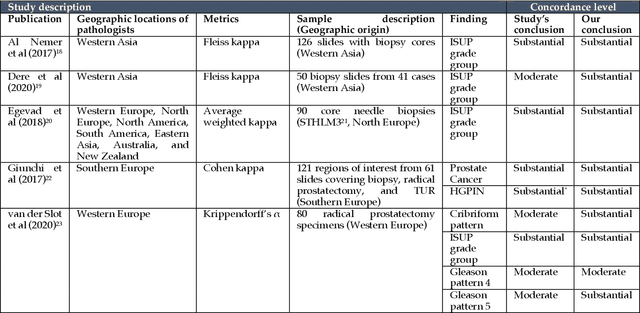
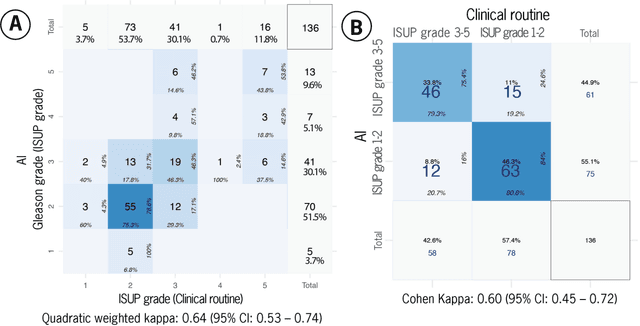
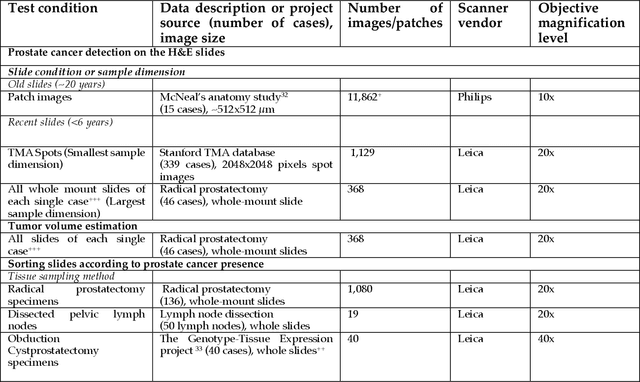
Abstract:Prostate cancer pathology plays a crucial role in clinical management but is time-consuming. Artificial intelligence (AI) shows promise in detecting prostate cancer and grading patterns. We tested an AI-based digital twin of a pathologist, vPatho, on 2,603 histology images of prostate tissue stained with hematoxylin and eosin. We analyzed various factors influencing tumor-grade disagreement between vPatho and six human pathologists. Our results demonstrated that vPatho achieved comparable performance in prostate cancer detection and tumor volume estimation, as reported in the literature. Concordance levels between vPatho and human pathologists were examined. Notably, moderate to substantial agreement was observed in identifying complementary histological features such as ductal, cribriform, nerve, blood vessels, and lymph cell infiltrations. However, concordance in tumor grading showed a decline when applied to prostatectomy specimens (kappa = 0.44) compared to biopsy cores (kappa = 0.70). Adjusting the decision threshold for the secondary Gleason pattern from 5% to 10% improved the concordance level between pathologists and vPatho for tumor grading on prostatectomy specimens (kappa from 0.44 to 0.64). Potential causes of grade discordance included the vertical extent of tumors toward the prostate boundary and the proportions of slides with prostate cancer. Gleason pattern 4 was particularly associated with discordance. Notably, grade discordance with vPatho was not specific to any of the six pathologists involved in routine clinical grading. In conclusion, our study highlights the potential utility of AI in developing a digital twin of a pathologist. This approach can help uncover limitations in AI adoption and the current grading system for prostate cancer pathology.
Collaborative Quantization Embeddings for Intra-Subject Prostate MR Image Registration
Jul 14, 2022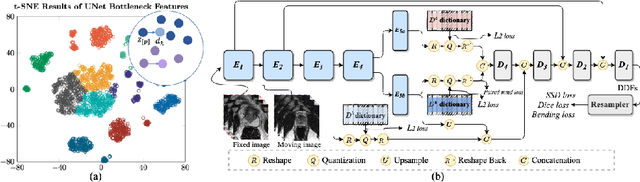
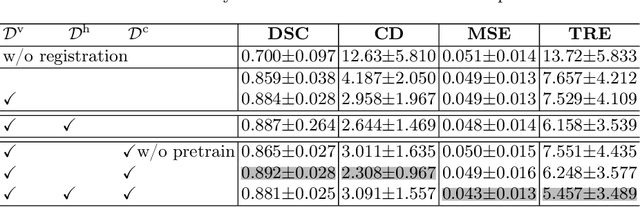
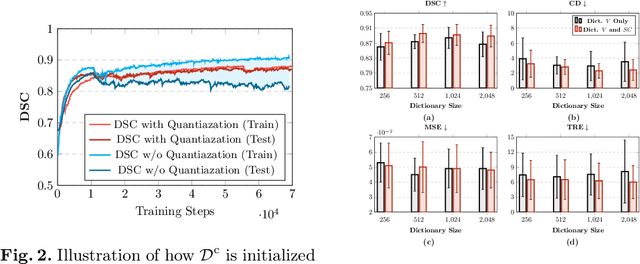
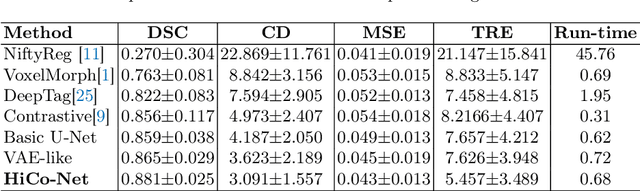
Abstract:Image registration is useful for quantifying morphological changes in longitudinal MR images from prostate cancer patients. This paper describes a development in improving the learning-based registration algorithms, for this challenging clinical application often with highly variable yet limited training data. First, we report that the latent space can be clustered into a much lower dimensional space than that commonly found as bottleneck features at the deep layer of a trained registration network. Based on this observation, we propose a hierarchical quantization method, discretizing the learned feature vectors using a jointly-trained dictionary with a constrained size, in order to improve the generalisation of the registration networks. Furthermore, a novel collaborative dictionary is independently optimised to incorporate additional prior information, such as the segmentation of the gland or other regions of interest, in the latent quantized space. Based on 216 real clinical images from 86 prostate cancer patients, we show the efficacy of both the designed components. Improved registration accuracy was obtained with statistical significance, in terms of both Dice on gland and target registration error on corresponding landmarks, the latter of which achieved 5.46 mm, an improvement of 28.7\% from the baseline without quantization. Experimental results also show that the difference in performance was indeed minimised between training and testing data.
Deep Learning for Prostate Pathology
Oct 16, 2019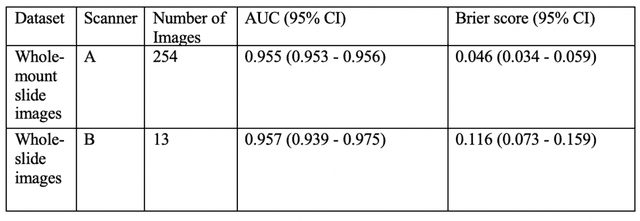
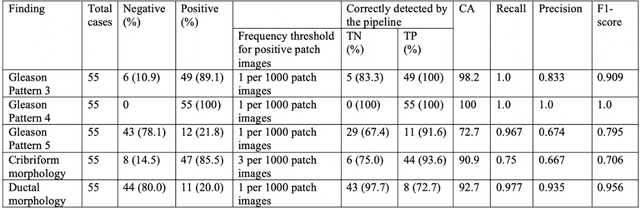
Abstract:The current study detects different morphologies related to prostate pathology using deep learning models; these models were evaluated on 2,121 hematoxylin and eosin (H&E) stain histology images captured using bright field microscopy, which spanned a variety of image qualities, origins (whole slide, tissue micro array, whole mount, Internet), scanning machines, timestamps, H&E staining protocols, and institutions. For case usage, these models were applied for the annotation tasks in clinician-oriented pathology reports for prostatectomy specimens. The true positive rate (TPR) for slides with prostate cancer was 99.7% by a false positive rate of 0.785%. The F1-scores of Gleason patterns reported in pathology reports ranged from 0.795 to 1.0 at the case level. TPR was 93.6% for the cribriform morphology and 72.6% for the ductal morphology. The correlation between the ground truth and the prediction for the relative tumor volume was 0.987 n. Our models cover the major components of prostate pathology and successfully accomplish the annotation tasks.
Computerized Multiparametric MR image Analysis for Prostate Cancer Aggressiveness-Assessment
Dec 01, 2016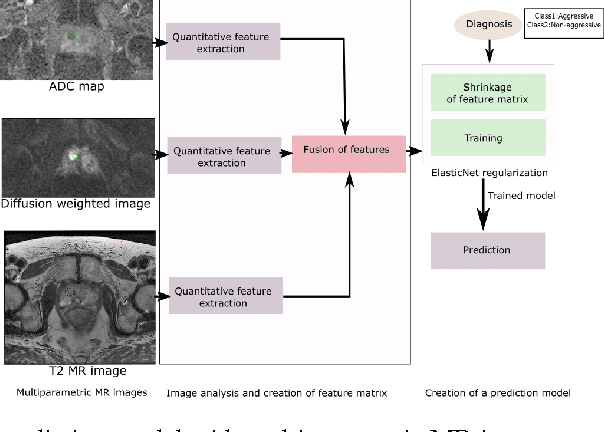
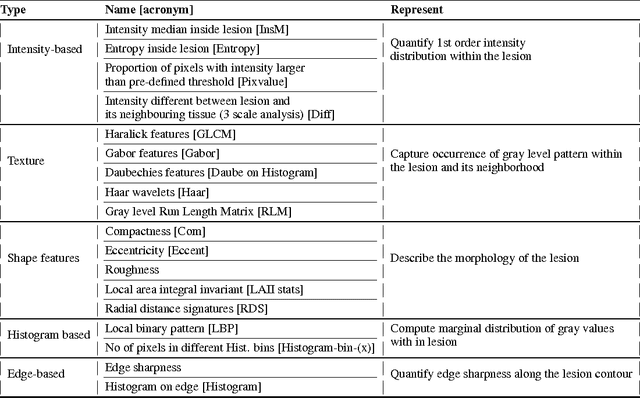
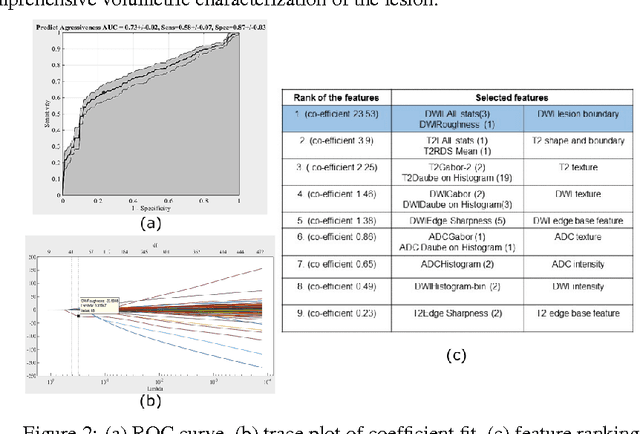
Abstract:We propose an automated method for detecting aggressive prostate cancer(CaP) (Gleason score >=7) based on a comprehensive analysis of the lesion and the surrounding normal prostate tissue which has been simultaneously captured in T2-weighted MR images, diffusion-weighted images (DWI) and apparent diffusion coefficient maps (ADC). The proposed methodology was tested on a dataset of 79 patients (40 aggressive, 39 non-aggressive). We evaluated the performance of a wide range of popular quantitative imaging features on the characterization of aggressive versus non-aggressive CaP. We found that a group of 44 discriminative predictors among 1464 quantitative imaging features can be used to produce an area under the ROC curve of 0.73.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge