Daniel L. Rubin
Spatiotemporal Modeling of Multivariate Signals With Graph Neural Networks and Structured State Space Models
Nov 21, 2022Abstract:Multivariate signals are prevalent in various domains, such as healthcare, transportation systems, and space sciences. Modeling spatiotemporal dependencies in multivariate signals is challenging due to (1) long-range temporal dependencies and (2) complex spatial correlations between sensors. To address these challenges, we propose representing multivariate signals as graphs and introduce GraphS4mer, a general graph neural network (GNN) architecture that captures both spatial and temporal dependencies in multivariate signals. Specifically, (1) we leverage Structured State Spaces model (S4), a state-of-the-art sequence model, to capture long-term temporal dependencies and (2) we propose a graph structure learning layer in GraphS4mer to learn dynamically evolving graph structures in the data. We evaluate our proposed model on three distinct tasks and show that GraphS4mer consistently improves over existing models, including (1) seizure detection from electroencephalography signals, outperforming a previous GNN with self-supervised pretraining by 3.1 points in AUROC; (2) sleep staging from polysomnography signals, a 4.1 points improvement in macro-F1 score compared to existing sleep staging models; and (3) traffic forecasting, reducing MAE by 8.8% compared to existing GNNs and by 1.4% compared to Transformer-based models.
TRUST-LAPSE: An Explainable & Actionable Mistrust Scoring Framework for Model Monitoring
Jul 22, 2022



Abstract:Continuous monitoring of trained ML models to determine when their predictions should and should not be trusted is essential for their safe deployment. Such a framework ought to be high-performing, explainable, post-hoc and actionable. We propose TRUST-LAPSE, a "mistrust" scoring framework for continuous model monitoring. We assess the trustworthiness of each input sample's model prediction using a sequence of latent-space embeddings. Specifically, (a) our latent-space mistrust score estimates mistrust using distance metrics (Mahalanobis distance) and similarity metrics (cosine similarity) in the latent-space and (b) our sequential mistrust score determines deviations in correlations over the sequence of past input representations in a non-parametric, sliding-window based algorithm for actionable continuous monitoring. We evaluate TRUST-LAPSE via two downstream tasks: (1) distributionally shifted input detection and (2) data drift detection, across diverse domains -- audio & vision using public datasets and further benchmark our approach on challenging, real-world electroencephalograms (EEG) datasets for seizure detection. Our latent-space mistrust scores achieve state-of-the-art results with AUROCs of 84.1 (vision), 73.9 (audio), 77.1 (clinical EEGs), outperforming baselines by over 10 points. We expose critical failures in popular baselines that remain insensitive to input semantic content, rendering them unfit for real-world model monitoring. We show that our sequential mistrust scores achieve high drift detection rates: over 90% of the streams show < 20% error for all domains. Through extensive qualitative and quantitative evaluations, we show that our mistrust scores are more robust and provide explainability for easy adoption into practice.
Supervised Machine Learning Algorithm for Detecting Consistency between Reported Findings and the Conclusions of Mammography Reports
Feb 28, 2022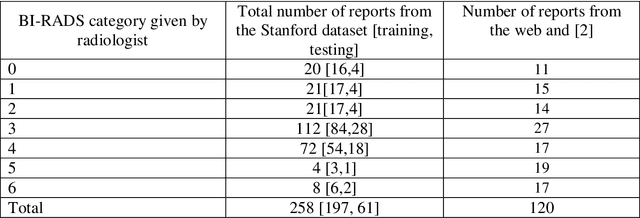
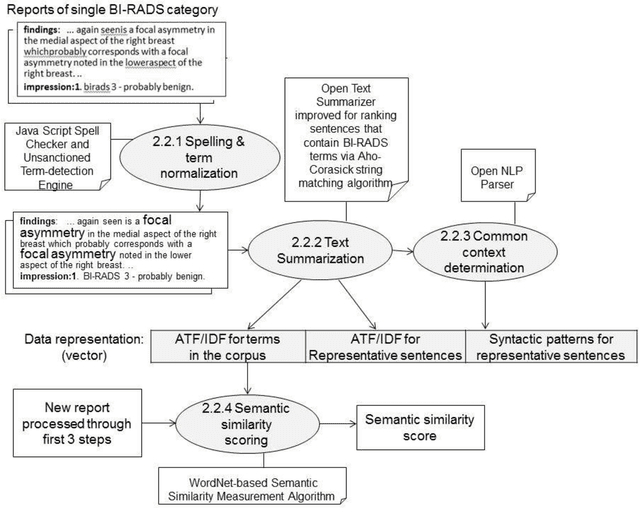
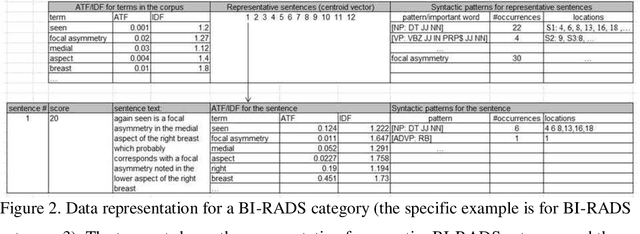
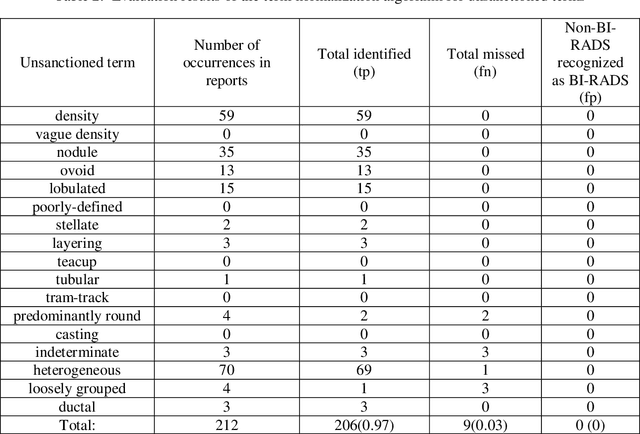
Abstract:Objective. Mammography reports document the diagnosis of patients' conditions. However, many reports contain non-standard terms (non-BI-RADS descriptors) and incomplete statements, which can lead to conclusions that are not well-supported by the reported findings. Our aim was to develop a tool to detect such discrepancies by comparing the reported conclusions to those that would be expected based on the reported radiology findings. Materials and Methods. A deidentified data set from an academic hospital containing 258 mammography reports supplemented by 120 reports found on the web was used for training and evaluation. Spell checking and term normalization was used to unambiguously determine the reported BI-RADS descriptors. The resulting data were input into seven classifiers that classify mammography reports, based on their Findings sections, into seven BI-RADS final assessment categories. Finally, the semantic similarity score of a report to each BI-RADS category is reported. Results. Our term normalization algorithm correctly identified 97% of the BI-RADS descriptors in mammography reports. Our system provided 76% precision and 83% recall in correctly classifying the reports according to BI-RADS final assessment category. Discussion. The strength of our approach relies on providing high importance to BI-RADS terms in the summarization phase, on the semantic similarity that considers the complex data representation, and on the classification into all seven BI-RADs categories. Conclusion. BI-RADS descriptors and expected final assessment categories could be automatically detected by our approach with fairly good accuracy, which could be used to make users aware that their reported findings do not match well with their conclusion.
SplitAVG: A heterogeneity-aware federated deep learning method for medical imaging
Jul 06, 2021



Abstract:Federated learning is an emerging research paradigm for enabling collaboratively training deep learning models without sharing patient data. However, the data from different institutions are usually heterogeneous across institutions, which may reduce the performance of models trained using federated learning. In this study, we propose a novel heterogeneity-aware federated learning method, SplitAVG, to overcome the performance drops from data heterogeneity in federated learning. Unlike previous federated methods that require complex heuristic training or hyper parameter tuning, our SplitAVG leverages the simple network split and feature map concatenation strategies to encourage the federated model training an unbiased estimator of the target data distribution. We compare SplitAVG with seven state-of-the-art federated learning methods, using centrally hosted training data as the baseline on a suite of both synthetic and real-world federated datasets. We find that the performance of models trained using all the comparison federated learning methods degraded significantly with the increasing degrees of data heterogeneity. In contrast, SplitAVG method achieves comparable results to the baseline method under all heterogeneous settings, that it achieves 96.2% of the accuracy and 110.4% of the mean absolute error obtained by the baseline in a diabetic retinopathy binary classification dataset and a bone age prediction dataset, respectively, on highly heterogeneous data partitions. We conclude that SplitAVG method can effectively overcome the performance drops from variability in data distributions across institutions. Experimental results also show that SplitAVG can be adapted to different base networks and generalized to various types of medical imaging tasks.
COVID-19 Lung Lesion Segmentation Using a Sparsely Supervised Mask R-CNN on Chest X-rays Automatically Computed from Volumetric CTs
May 20, 2021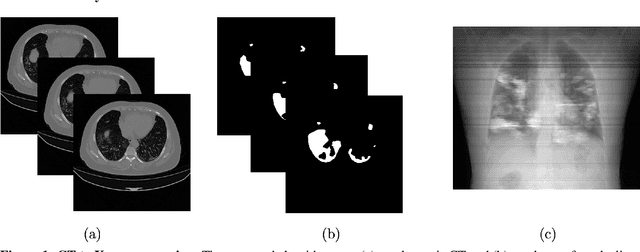
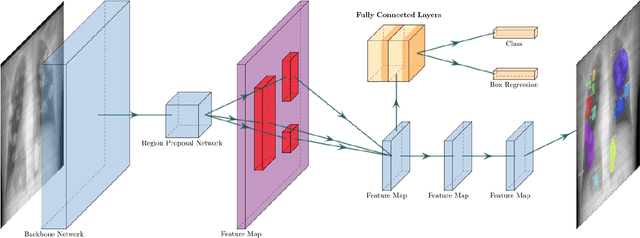
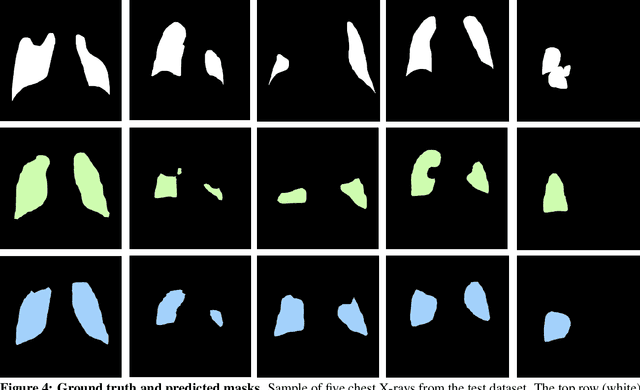
Abstract:Chest X-rays of coronavirus disease 2019 (COVID-19) patients are frequently obtained to determine the extent of lung disease and are a valuable source of data for creating artificial intelligence models. Most work to date assessing disease severity on chest imaging has focused on segmenting computed tomography (CT) images; however, given that CTs are performed much less frequently than chest X-rays for COVID-19 patients, automated lung lesion segmentation on chest X-rays could be clinically valuable. There currently exists a universal shortage of chest X-rays with ground truth COVID-19 lung lesion annotations, and manually contouring lung opacities is a tedious, labor-intensive task. To accelerate severity detection and augment the amount of publicly available chest X-ray training data for supervised deep learning (DL) models, we leverage existing annotated CT images to generate frontal projection "chest X-ray" images for training COVID-19 chest X-ray models. In this paper, we propose an automated pipeline for segmentation of COVID-19 lung lesions on chest X-rays comprised of a Mask R-CNN trained on a mixed dataset of open-source chest X-rays and coronal X-ray projections computed from annotated volumetric CTs. On a test set containing 40 chest X-rays of COVID-19 positive patients, our model achieved IoU scores of 0.81 $\pm$ 0.03 and 0.79 $\pm$ 0.03 when trained on a dataset of 60 chest X-rays and on a mixed dataset of 10 chest X-rays and 50 projections from CTs, respectively. Our model far outperforms current baselines with limited supervised training and may assist in automated COVID-19 severity quantification on chest X-rays.
Automated Seizure Detection and Seizure Type Classification From Electroencephalography With a Graph Neural Network and Self-Supervised Pre-Training
Apr 16, 2021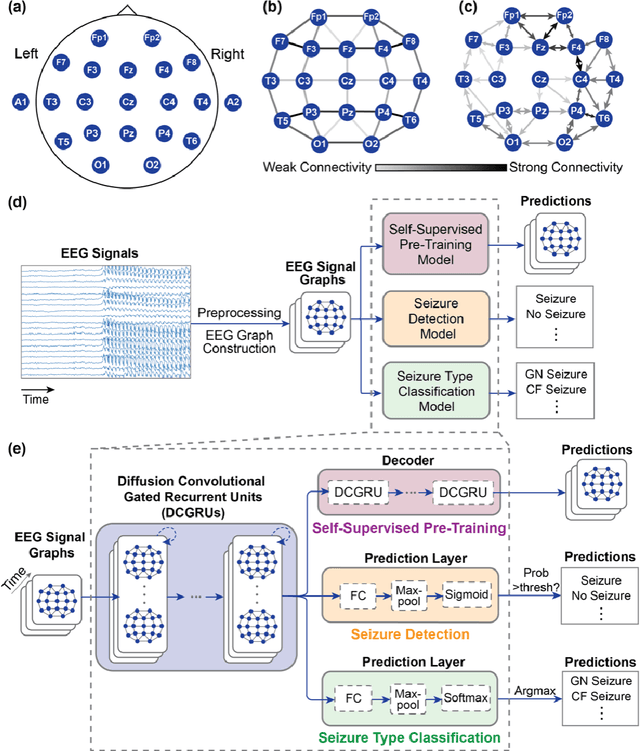
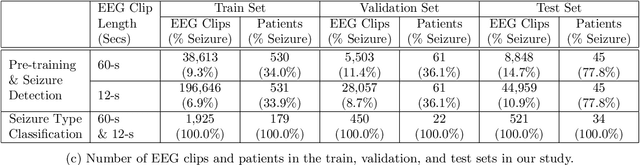
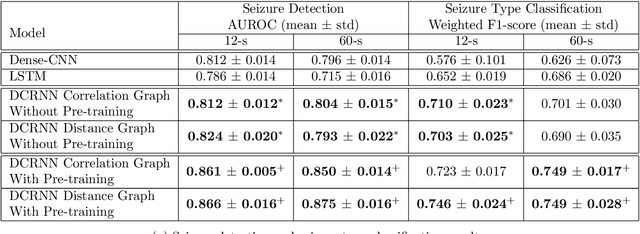
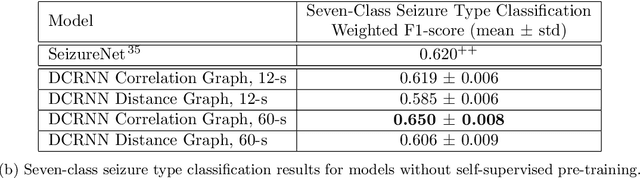
Abstract:Automated seizure detection and classification from electroencephalography (EEG) can greatly improve the diagnosis and treatment of seizures. While prior studies mainly used convolutional neural networks (CNNs) that assume image-like structure in EEG signals or spectrograms, this modeling choice does not reflect the natural geometry of or connectivity between EEG electrodes. In this study, we propose modeling EEGs as graphs and present a graph neural network for automated seizure detection and classification. In addition, we leverage unlabeled EEG data using a self-supervised pre-training strategy. Our graph model with self-supervised pre-training significantly outperforms previous state-of-the-art CNN and Long Short-Term Memory (LSTM) models by 6.3 points (7.8%) in Area Under the Receiver Operating Characteristic curve (AUROC) for seizure detection and 6.3 points (9.2%) in weighted F1-score for seizure type classification. Ablation studies show that our graph-based modeling approach significantly outperforms existing CNN or LSTM models, and that self-supervision helps further improve the model performance. Moreover, we find that self-supervised pre-training substantially improves model performance on combined tonic seizures, a low-prevalence seizure type. Furthermore, our model interpretability analysis suggests that our model is better at identifying seizure regions compared to an existing CNN. In summary, our graph-based modeling approach integrates domain knowledge about EEG, sets a new state-of-the-art for seizure detection and classification on a large public dataset (5,499 EEG files), and provides better ability to identify seizure regions.
Addressing catastrophic forgetting for medical domain expansion
Mar 24, 2021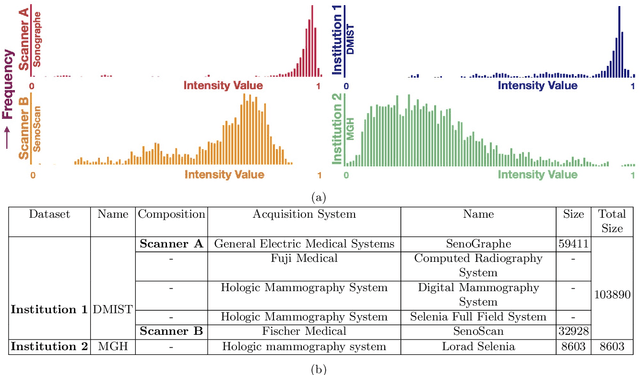
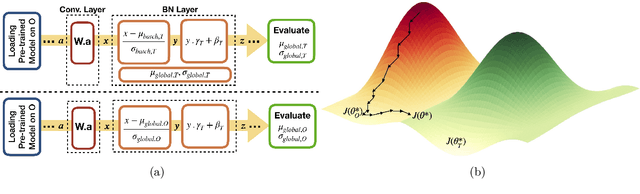
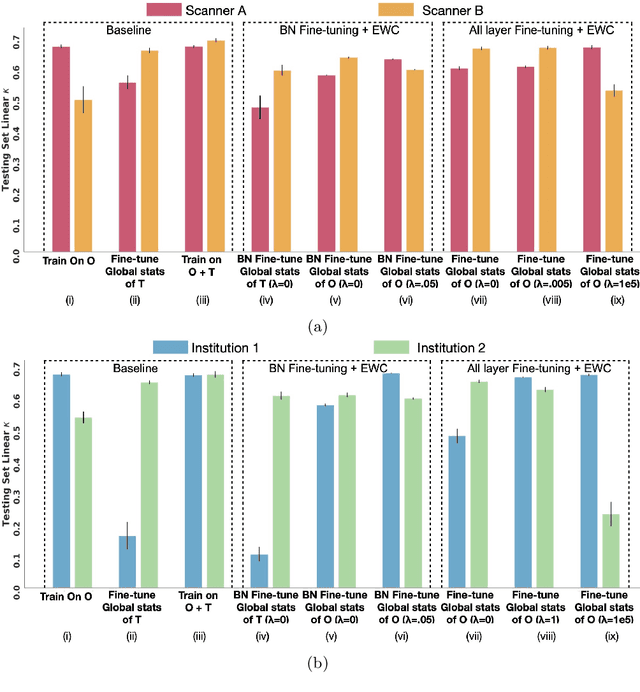
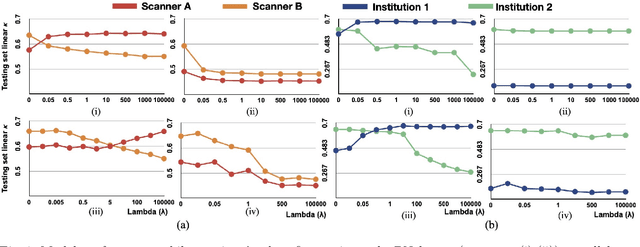
Abstract:Model brittleness is a key concern when deploying deep learning models in real-world medical settings. A model that has high performance at one institution may suffer a significant decline in performance when tested at other institutions. While pooling datasets from multiple institutions and retraining may provide a straightforward solution, it is often infeasible and may compromise patient privacy. An alternative approach is to fine-tune the model on subsequent institutions after training on the original institution. Notably, this approach degrades model performance at the original institution, a phenomenon known as catastrophic forgetting. In this paper, we develop an approach to address catastrophic forget-ting based on elastic weight consolidation combined with modulation of batch normalization statistics under two scenarios: first, for expanding the domain from one imaging system's data to another imaging system's, and second, for expanding the domain from a large multi-institutional dataset to another single institution dataset. We show that our approach outperforms several other state-of-the-art approaches and provide theoretical justification for the efficacy of batch normalization modulation. The results of this study are generally applicable to the deployment of any clinical deep learning model which requires domain expansion.
Learning domain-agnostic visual representation for computational pathology using medically-irrelevant style transfer augmentation
Feb 02, 2021



Abstract:Suboptimal generalization of machine learning models on unseen data is a key challenge which hampers the clinical applicability of such models to medical imaging. Although various methods such as domain adaptation and domain generalization have evolved to combat this challenge, learning robust and generalizable representations is core to medical image understanding, and continues to be a problem. Here, we propose STRAP (Style TRansfer Augmentation for histoPathology), a form of data augmentation based on random style transfer from artistic paintings, for learning domain-agnostic visual representations in computational pathology. Style transfer replaces the low-level texture content of images with the uninformative style of randomly selected artistic paintings, while preserving high-level semantic content. This improves robustness to domain shift and can be used as a simple yet powerful tool for learning domain-agnostic representations. We demonstrate that STRAP leads to state-of-the-art performance, particularly in the presence of domain shifts, on a particular classification task of predicting microsatellite status in colorectal cancer using digitized histopathology images.
Data Valuation for Medical Imaging Using Shapley Value: Application on A Large-scale Chest X-ray Dataset
Oct 15, 2020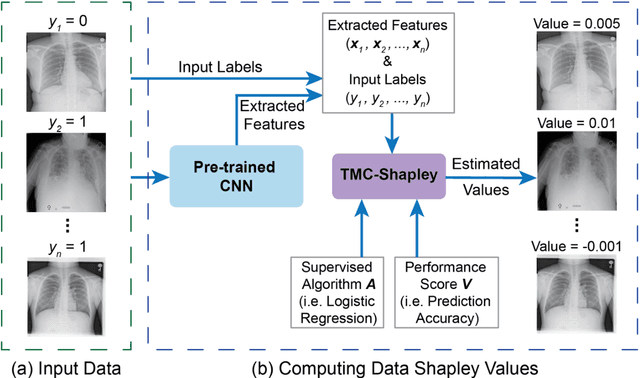
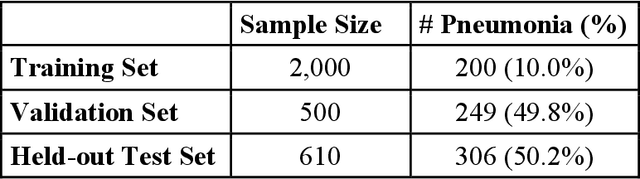
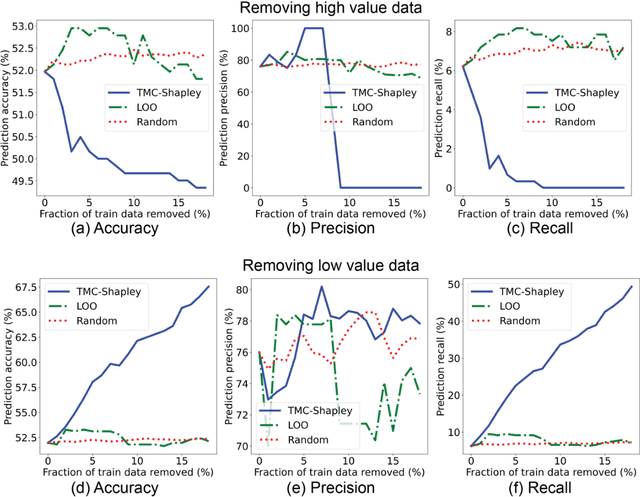

Abstract:The reliability of machine learning models can be compromised when trained on low quality data. Many large-scale medical imaging datasets contain low quality labels extracted from sources such as medical reports. Moreover, images within a dataset may have heterogeneous quality due to artifacts and biases arising from equipment or measurement errors. Therefore, algorithms that can automatically identify low quality data are highly desired. In this study, we used data Shapley, a data valuation metric, to quantify the value of training data to the performance of a pneumonia detection algorithm in a large chest X-ray dataset. We characterized the effectiveness of data Shapley in identifying low quality versus valuable data for pneumonia detection. We found that removing training data with high Shapley values decreased the pneumonia detection performance, whereas removing data with low Shapley values improved the model performance. Furthermore, there were more mislabeled examples in low Shapley value data and more true pneumonia cases in high Shapley value data. Our results suggest that low Shapley value indicates mislabeled or poor quality images, whereas high Shapley value indicates data that are valuable for pneumonia detection. Our method can serve as a framework for using data Shapley to denoise large-scale medical imaging datasets.
Probabilistic bounds on data sensitivity in deep rectifier networks
Jul 13, 2020

Abstract:Neuron death is a complex phenomenon with implications for model trainability, but until recently it was measured only empirically. Recent articles have claimed that, as the depth of a rectifier neural network grows to infinity, the probability of finding a valid initialization decreases to zero. In this work, we provide a simple and rigorous proof of that result. Then, we show what happens when the width of each layer grows simultaneously with the depth. We derive both upper and lower bounds on the probability that a ReLU network is initialized to a trainable point, as a function of model hyperparameters. Contrary to previous claims, we show that it is possible to increase the depth of a network indefinitely, so long as the width increases as well. Furthermore, our bounds are asymptotically tight under reasonable assumptions: first, the upper bound coincides with the true probability for a single-layer network with the largest possible input set. Second, the true probability converges to our lower bound when the network width and depth both grow without limit. Our proof is based on the striking observation that very deep rectifier networks concentrate all outputs towards a single eigenvalue, in the sense that their normalized output variance goes to zero regardless of the network width. Finally, we develop a practical sign flipping scheme which guarantees with probability one that for a $k$-layer network, the ratio of living training data points is at least $2^{-k}$. We confirm our results with numerical simulations, suggesting that the actual improvement far exceeds the theoretical minimum. We also discuss how neuron death provides a theoretical interpretation for various network design choices such as batch normalization, residual layers and skip connections, and could inform the design of very deep neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge