Vignav Ramesh
Communicating Activations Between Language Model Agents
Jan 23, 2025Abstract:Communication between multiple language model (LM) agents has been shown to scale up the reasoning ability of LMs. While natural language has been the dominant medium for inter-LM communication, it is not obvious this should be the standard: not only does natural language communication incur high inference costs that scale quickly with the number of both agents and messages, but also the decoding process abstracts away too much rich information that could be otherwise accessed from the internal activations. In this work, we propose a simple technique whereby LMs communicate via activations; concretely, we pause an LM $\textit{B}$'s computation at an intermediate layer, combine its current activation with another LM $\textit{A}$'s intermediate activation via some function $\textit{f}$, then pass $\textit{f}$'s output into the next layer of $\textit{B}$ and continue the forward pass till decoding is complete. This approach scales up LMs on new tasks with zero additional parameters and data, and saves a substantial amount of compute over natural language communication. We test our method with various functional forms $\textit{f}$ on two experimental setups--multi-player coordination games and reasoning benchmarks--and find that it achieves up to $27.0\%$ improvement over natural language communication across datasets with $<$$1/4$ the compute, illustrating the superiority and robustness of activations as an alternative "language" for communication between LMs.
Improving Radiology Report Generation Systems by Removing Hallucinated References to Non-existent Priors
Sep 27, 2022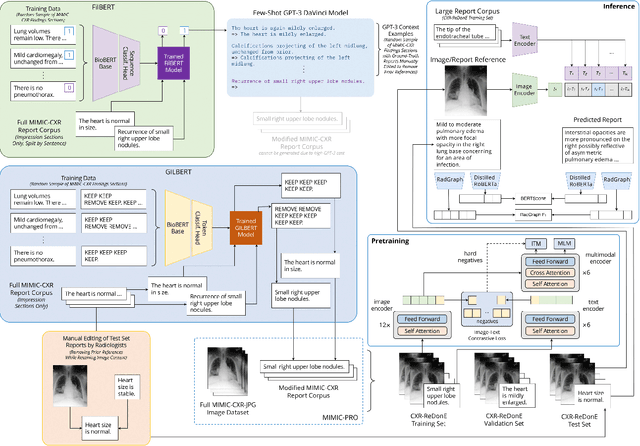
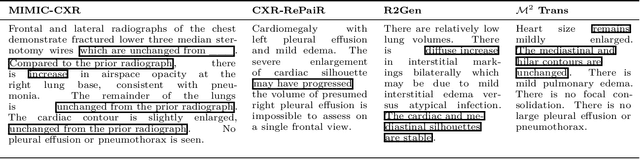
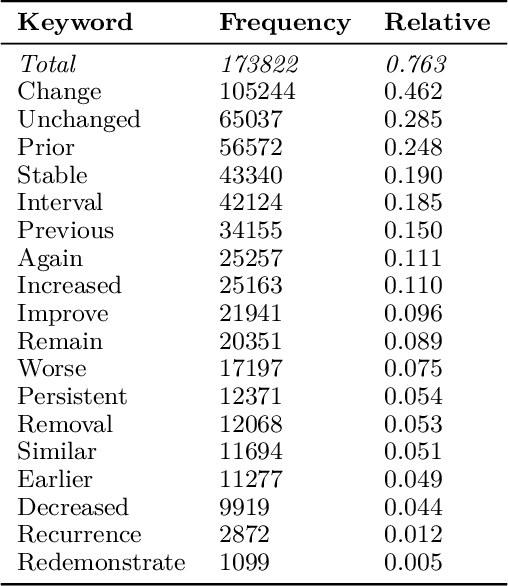
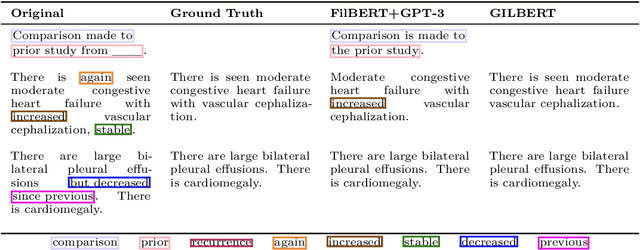
Abstract:Current deep learning models trained to generate radiology reports from chest radiographs are capable of producing clinically accurate, clear, and actionable text that can advance patient care. However, such systems all succumb to the same problem: making hallucinated references to non-existent prior reports. Such hallucinations occur because these models are trained on datasets of real-world patient reports that inherently refer to priors. To this end, we propose two methods to remove references to priors in radiology reports: (1) a GPT-3-based few-shot approach to rewrite medical reports without references to priors; and (2) a BioBERT-based token classification approach to directly remove words referring to priors. We use the aforementioned approaches to modify MIMIC-CXR, a publicly available dataset of chest X-rays and their associated free-text radiology reports; we then retrain CXR-RePaiR, a radiology report generation system, on the adapted MIMIC-CXR dataset. We find that our re-trained model--which we call CXR-ReDonE--outperforms previous report generation methods on clinical metrics, achieving an average BERTScore of 0.2351 (2.57% absolute improvement). We expect our approach to be broadly valuable in enabling current radiology report generation systems to be more directly integrated into clinical pipelines.
Contrastive learning-based pretraining improves representation and transferability of diabetic retinopathy classification models
Aug 24, 2022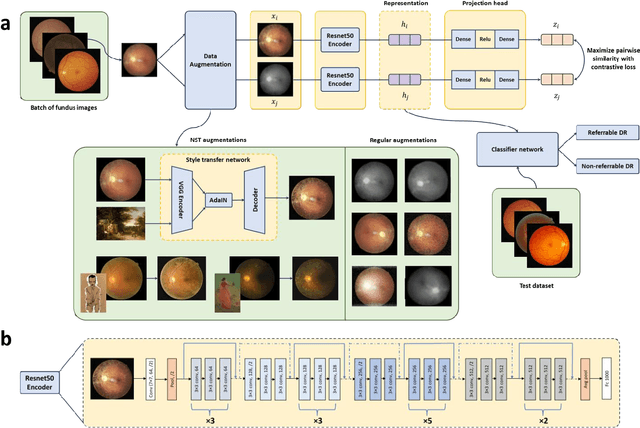
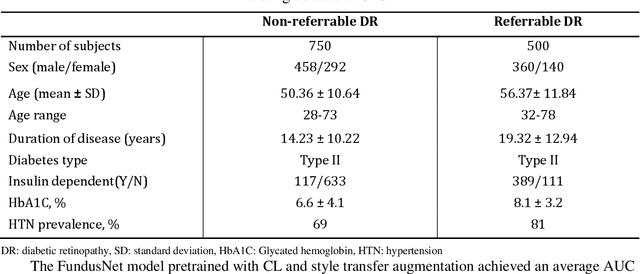
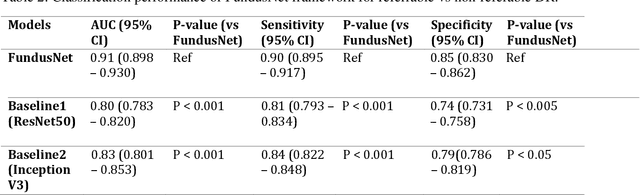
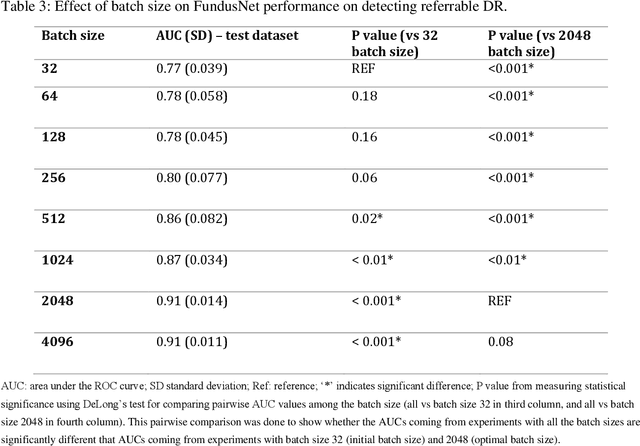
Abstract:Self supervised contrastive learning based pretraining allows development of robust and generalized deep learning models with small, labeled datasets, reducing the burden of label generation. This paper aims to evaluate the effect of CL based pretraining on the performance of referrable vs non referrable diabetic retinopathy (DR) classification. We have developed a CL based framework with neural style transfer (NST) augmentation to produce models with better representations and initializations for the detection of DR in color fundus images. We compare our CL pretrained model performance with two state of the art baseline models pretrained with Imagenet weights. We further investigate the model performance with reduced labeled training data (down to 10 percent) to test the robustness of the model when trained with small, labeled datasets. The model is trained and validated on the EyePACS dataset and tested independently on clinical data from the University of Illinois, Chicago (UIC). Compared to baseline models, our CL pretrained FundusNet model had higher AUC (CI) values (0.91 (0.898 to 0.930) vs 0.80 (0.783 to 0.820) and 0.83 (0.801 to 0.853) on UIC data). At 10 percent labeled training data, the FundusNet AUC was 0.81 (0.78 to 0.84) vs 0.58 (0.56 to 0.64) and 0.63 (0.60 to 0.66) in baseline models, when tested on the UIC dataset. CL based pretraining with NST significantly improves DL classification performance, helps the model generalize well (transferable from EyePACS to UIC data), and allows training with small, annotated datasets, therefore reducing ground truth annotation burden of the clinicians.
End-to-end Malaria Diagnosis and 3D Cell Rendering with Deep Learning
Jul 08, 2021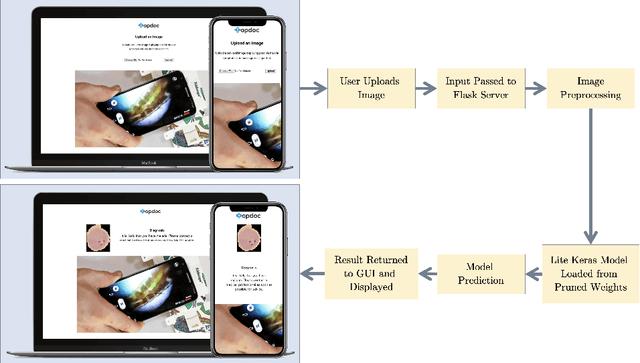
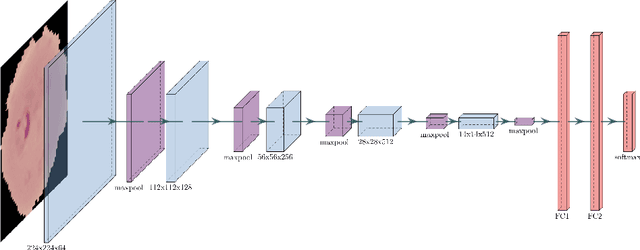
Abstract:Malaria is a parasitic infection that poses a significant burden on global health. It kills one child every 30 seconds and over one million people annually. If diagnosed in a timely manner, however, most people can be effectively treated with antimalarial therapy. Several deaths due to malaria are byproducts of disparities in the social determinants of health; the current gold standard for diagnosing malaria requires microscopes, reagents, and other equipment that most patients of low socioeconomic brackets do not have access to. In this paper, we propose a convolutional neural network (CNN) architecture that allows for rapid automated diagnosis of malaria (achieving a high classification accuracy of 98%), as well as a deep neural network (DNN) based three-dimensional (3D) modeling algorithm that renders 3D models of parasitic cells in augmented reality (AR). This creates an opportunity to optimize the current workflow for malaria diagnosis and demonstrates potential for deep learning models to improve telemedicine practices and patient health literacy on a global scale.
COVID-19 Lung Lesion Segmentation Using a Sparsely Supervised Mask R-CNN on Chest X-rays Automatically Computed from Volumetric CTs
May 20, 2021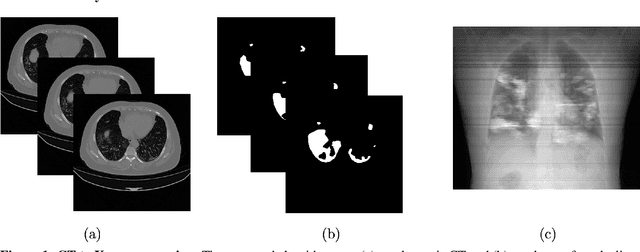
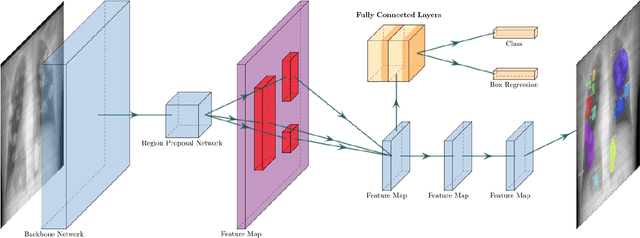
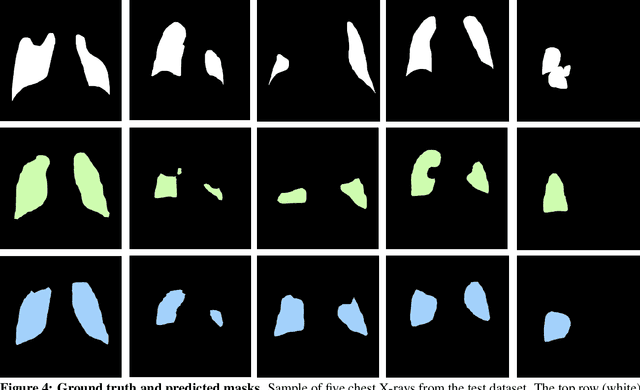
Abstract:Chest X-rays of coronavirus disease 2019 (COVID-19) patients are frequently obtained to determine the extent of lung disease and are a valuable source of data for creating artificial intelligence models. Most work to date assessing disease severity on chest imaging has focused on segmenting computed tomography (CT) images; however, given that CTs are performed much less frequently than chest X-rays for COVID-19 patients, automated lung lesion segmentation on chest X-rays could be clinically valuable. There currently exists a universal shortage of chest X-rays with ground truth COVID-19 lung lesion annotations, and manually contouring lung opacities is a tedious, labor-intensive task. To accelerate severity detection and augment the amount of publicly available chest X-ray training data for supervised deep learning (DL) models, we leverage existing annotated CT images to generate frontal projection "chest X-ray" images for training COVID-19 chest X-ray models. In this paper, we propose an automated pipeline for segmentation of COVID-19 lung lesions on chest X-rays comprised of a Mask R-CNN trained on a mixed dataset of open-source chest X-rays and coronal X-ray projections computed from annotated volumetric CTs. On a test set containing 40 chest X-rays of COVID-19 positive patients, our model achieved IoU scores of 0.81 $\pm$ 0.03 and 0.79 $\pm$ 0.03 when trained on a dataset of 60 chest X-rays and on a mixed dataset of 10 chest X-rays and 50 projections from CTs, respectively. Our model far outperforms current baselines with limited supervised training and may assist in automated COVID-19 severity quantification on chest X-rays.
Natural Language Generation Using Link Grammar for General Conversational Intelligence
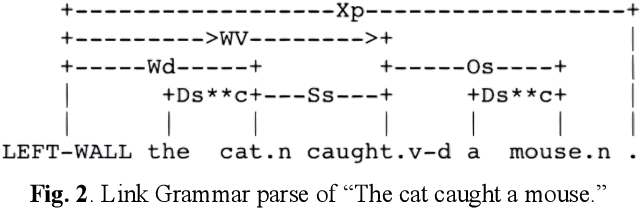
Apr 19, 2021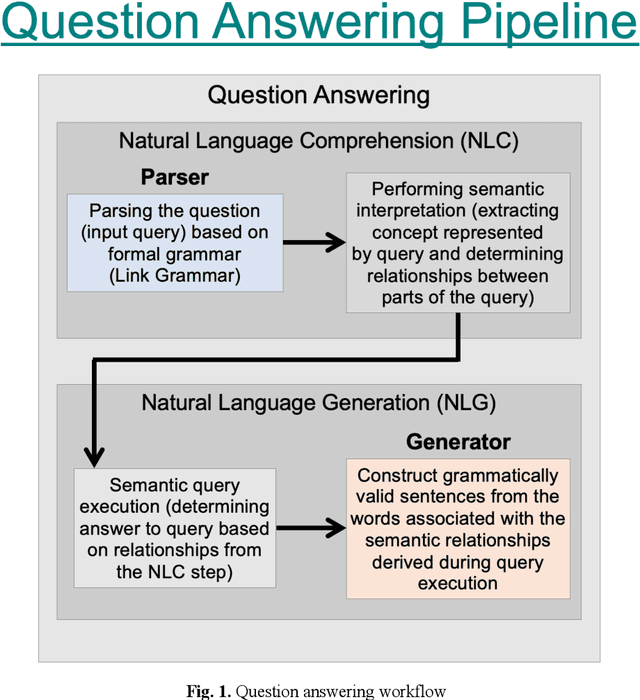
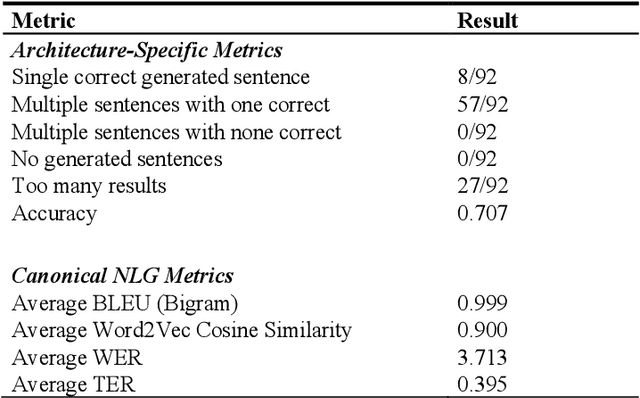
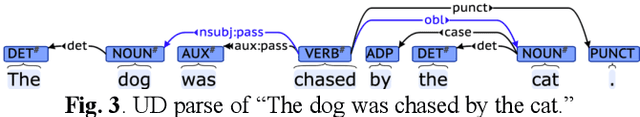
Abstract:Many current artificial general intelligence (AGI) and natural language processing (NLP) architectures do not possess general conversational intelligence--that is, they either do not deal with language or are unable to convey knowledge in a form similar to the human language without manual, labor-intensive methods such as template-based customization. In this paper, we propose a new technique to automatically generate grammatically valid sentences using the Link Grammar database. This natural language generation method far outperforms current state-of-the-art baselines and may serve as the final component in a proto-AGI question answering pipeline that understandably handles natural language material.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge