Rikiya Yamashita
Improved Multimodal Fusion for Small Datasets with Auxiliary Supervision
Apr 01, 2023Abstract:Prostate cancer is one of the leading causes of cancer-related death in men worldwide. Like many cancers, diagnosis involves expert integration of heterogeneous patient information such as imaging, clinical risk factors, and more. For this reason, there have been many recent efforts toward deep multimodal fusion of image and non-image data for clinical decision tasks. Many of these studies propose methods to fuse learned features from each patient modality, providing significant downstream improvements with techniques like cross-modal attention gating, Kronecker product fusion, orthogonality regularization, and more. While these enhanced fusion operations can improve upon feature concatenation, they often come with an extremely high learning capacity, meaning they are likely to overfit when applied even to small or low-dimensional datasets. Rather than designing a highly expressive fusion operation, we propose three simple methods for improved multimodal fusion with small datasets that aid optimization by generating auxiliary sources of supervision during training: extra supervision, clinical prediction, and dense fusion. We validate the proposed approaches on prostate cancer diagnosis from paired histopathology imaging and tabular clinical features. The proposed methods are straightforward to implement and can be applied to any classification task with paired image and non-image data.
Contrastive learning-based pretraining improves representation and transferability of diabetic retinopathy classification models
Aug 24, 2022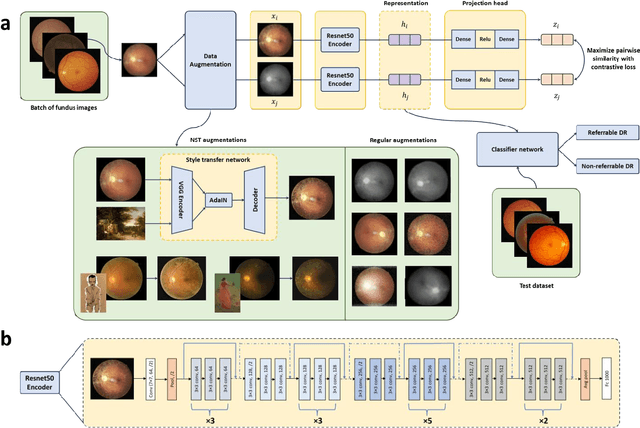
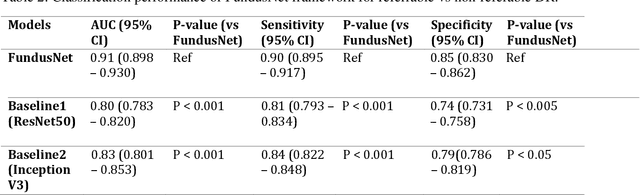
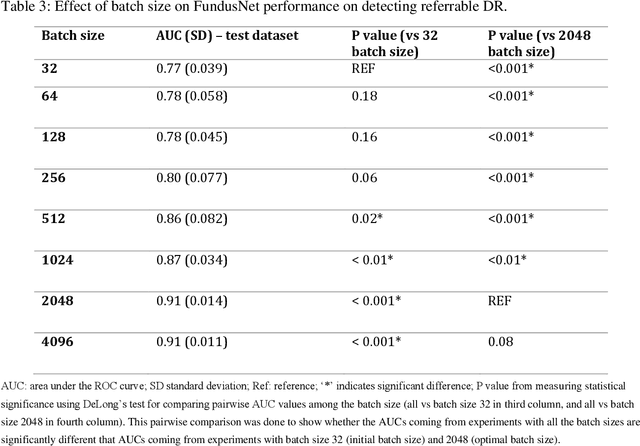
Abstract:Self supervised contrastive learning based pretraining allows development of robust and generalized deep learning models with small, labeled datasets, reducing the burden of label generation. This paper aims to evaluate the effect of CL based pretraining on the performance of referrable vs non referrable diabetic retinopathy (DR) classification. We have developed a CL based framework with neural style transfer (NST) augmentation to produce models with better representations and initializations for the detection of DR in color fundus images. We compare our CL pretrained model performance with two state of the art baseline models pretrained with Imagenet weights. We further investigate the model performance with reduced labeled training data (down to 10 percent) to test the robustness of the model when trained with small, labeled datasets. The model is trained and validated on the EyePACS dataset and tested independently on clinical data from the University of Illinois, Chicago (UIC). Compared to baseline models, our CL pretrained FundusNet model had higher AUC (CI) values (0.91 (0.898 to 0.930) vs 0.80 (0.783 to 0.820) and 0.83 (0.801 to 0.853) on UIC data). At 10 percent labeled training data, the FundusNet AUC was 0.81 (0.78 to 0.84) vs 0.58 (0.56 to 0.64) and 0.63 (0.60 to 0.66) in baseline models, when tested on the UIC dataset. CL based pretraining with NST significantly improves DL classification performance, helps the model generalize well (transferable from EyePACS to UIC data), and allows training with small, annotated datasets, therefore reducing ground truth annotation burden of the clinicians.
Learning domain-agnostic visual representation for computational pathology using medically-irrelevant style transfer augmentation
Feb 02, 2021



Abstract:Suboptimal generalization of machine learning models on unseen data is a key challenge which hampers the clinical applicability of such models to medical imaging. Although various methods such as domain adaptation and domain generalization have evolved to combat this challenge, learning robust and generalizable representations is core to medical image understanding, and continues to be a problem. Here, we propose STRAP (Style TRansfer Augmentation for histoPathology), a form of data augmentation based on random style transfer from artistic paintings, for learning domain-agnostic visual representations in computational pathology. Style transfer replaces the low-level texture content of images with the uninformative style of randomly selected artistic paintings, while preserving high-level semantic content. This improves robustness to domain shift and can be used as a simple yet powerful tool for learning domain-agnostic representations. We demonstrate that STRAP leads to state-of-the-art performance, particularly in the presence of domain shifts, on a particular classification task of predicting microsatellite status in colorectal cancer using digitized histopathology images.
Data Valuation for Medical Imaging Using Shapley Value: Application on A Large-scale Chest X-ray Dataset
Oct 15, 2020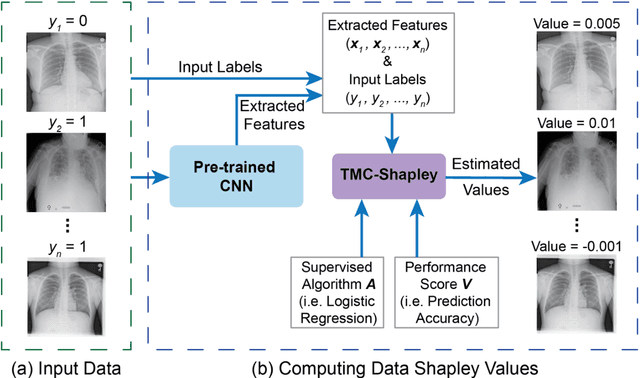
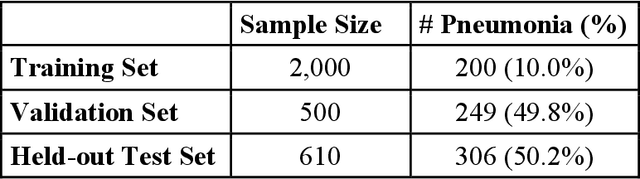
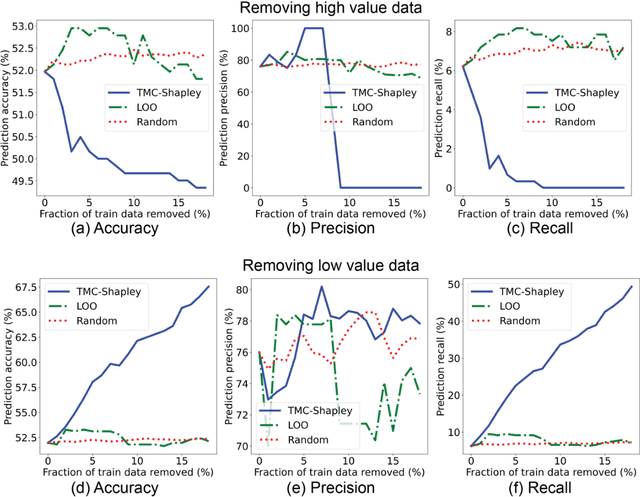

Abstract:The reliability of machine learning models can be compromised when trained on low quality data. Many large-scale medical imaging datasets contain low quality labels extracted from sources such as medical reports. Moreover, images within a dataset may have heterogeneous quality due to artifacts and biases arising from equipment or measurement errors. Therefore, algorithms that can automatically identify low quality data are highly desired. In this study, we used data Shapley, a data valuation metric, to quantify the value of training data to the performance of a pneumonia detection algorithm in a large chest X-ray dataset. We characterized the effectiveness of data Shapley in identifying low quality versus valuable data for pneumonia detection. We found that removing training data with high Shapley values decreased the pneumonia detection performance, whereas removing data with low Shapley values improved the model performance. Furthermore, there were more mislabeled examples in low Shapley value data and more true pneumonia cases in high Shapley value data. Our results suggest that low Shapley value indicates mislabeled or poor quality images, whereas high Shapley value indicates data that are valuable for pneumonia detection. Our method can serve as a framework for using data Shapley to denoise large-scale medical imaging datasets.
Analysis Of Multi Field Of View Cnn And Attention Cnn On H&E Stained Whole-slide Images On Hepatocellular Carcinoma
Feb 18, 2020



Abstract:Hepatocellular carcinoma (HCC) is a leading cause of cancer-related death worldwide. Whole-slide imaging which is a method of scanning glass slides have been employed for diagnosis of HCC. Using high resolution Whole-slide images is infeasible for Convolutional Neural Network applications. Hence tiling the Whole-slide images is a common methodology for assigning Convolutional Neural Networks for classification and segmentation. Determination of the tile size affects the performance of the algorithms since small field of view can not capture the information on a larger scale and large field of view can not capture the information on a cellular scale. In this work, the effect of tile size on performance for classification problem is analysed. In addition, Multi Field of View CNN is assigned for taking advantage of the information provided by different tile sizes and Attention CNN is assigned for giving the capability of voting most contributing tile size. It is found that employing more than one tile size significantly increases the performance of the classification by 3.97% and both algorithms are found successful over the algorithm which uses only one tile size.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge