Amirata Ghorbani
DataPerf: Benchmarks for Data-Centric AI Development
Jul 20, 2022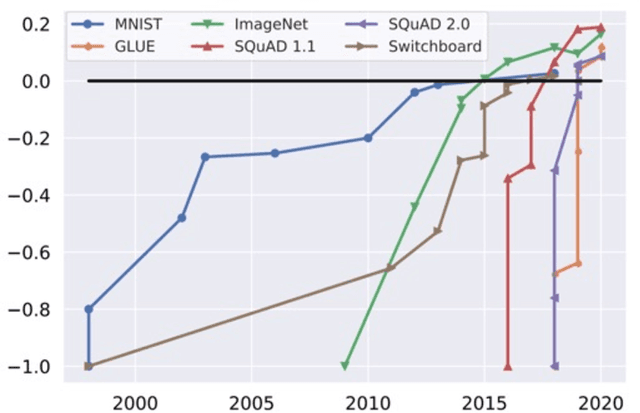
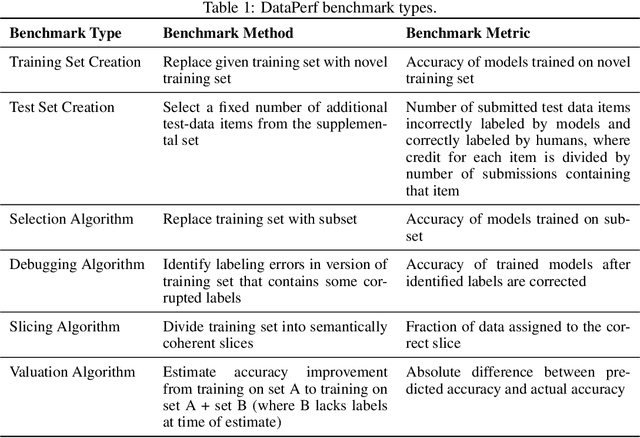
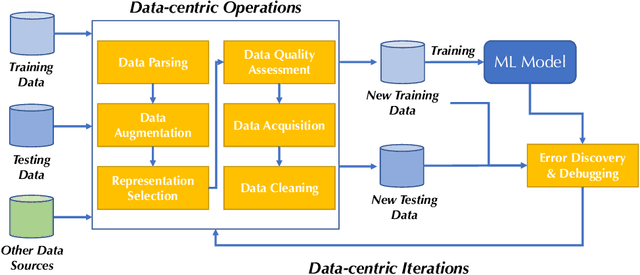
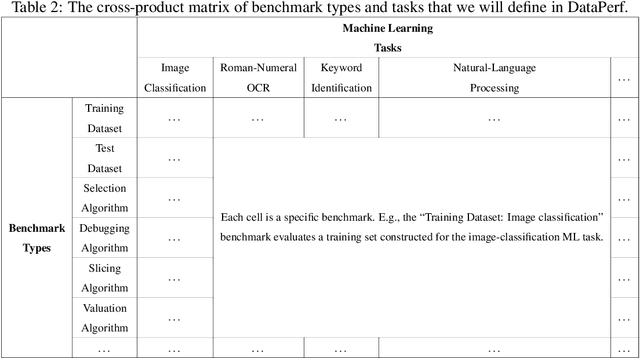
Abstract:Machine learning (ML) research has generally focused on models, while the most prominent datasets have been employed for everyday ML tasks without regard for the breadth, difficulty, and faithfulness of these datasets to the underlying problem. Neglecting the fundamental importance of datasets has caused major problems involving data cascades in real-world applications and saturation of dataset-driven criteria for model quality, hindering research growth. To solve this problem, we present DataPerf, a benchmark package for evaluating ML datasets and dataset-working algorithms. We intend it to enable the "data ratchet," in which training sets will aid in evaluating test sets on the same problems, and vice versa. Such a feedback-driven strategy will generate a virtuous loop that will accelerate development of data-centric AI. The MLCommons Association will maintain DataPerf.
Beyond Importance Scores: Interpreting Tabular ML by Visualizing Feature Semantics
Nov 30, 2021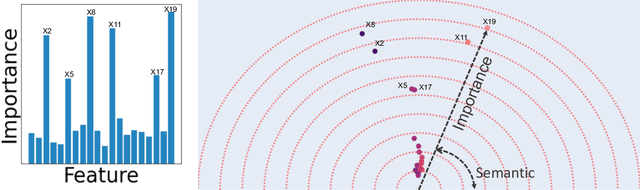
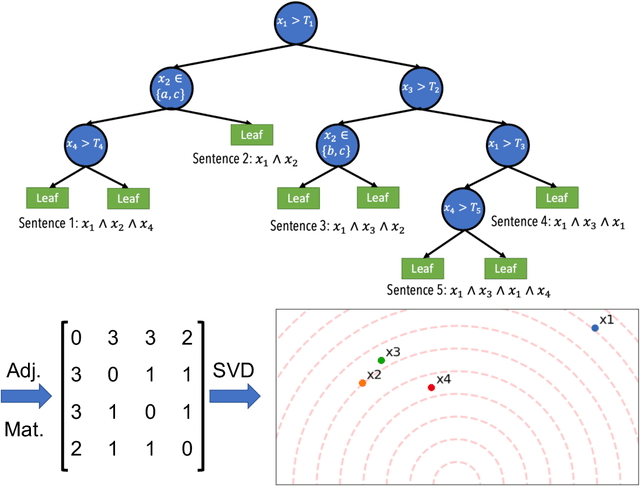
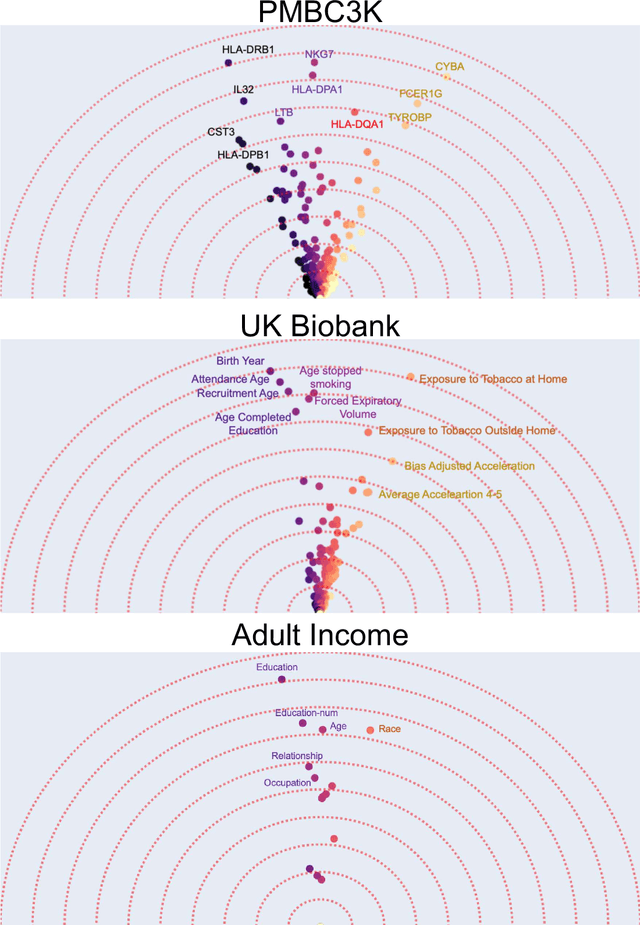
Abstract:Interpretability is becoming an active research topic as machine learning (ML) models are more widely used to make critical decisions. Tabular data is one of the most commonly used modes of data in diverse applications such as healthcare and finance. Much of the existing interpretability methods used for tabular data only report feature-importance scores -- either locally (per example) or globally (per model) -- but they do not provide interpretation or visualization of how the features interact. We address this limitation by introducing Feature Vectors, a new global interpretability method designed for tabular datasets. In addition to providing feature-importance, Feature Vectors discovers the inherent semantic relationship among features via an intuitive feature visualization technique. Our systematic experiments demonstrate the empirical utility of this new method by applying it to several real-world datasets. We further provide an easy-to-use Python package for Feature Vectors.
Data Shapley Valuation for Efficient Batch Active Learning
Apr 16, 2021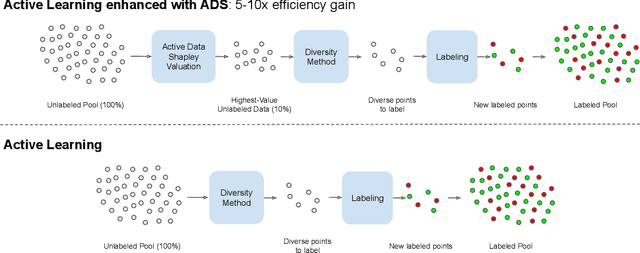
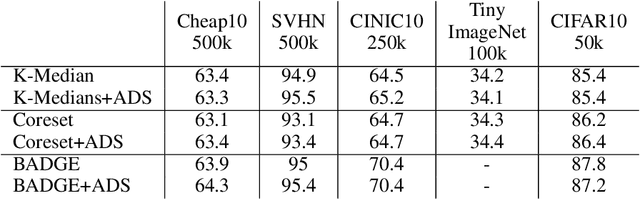
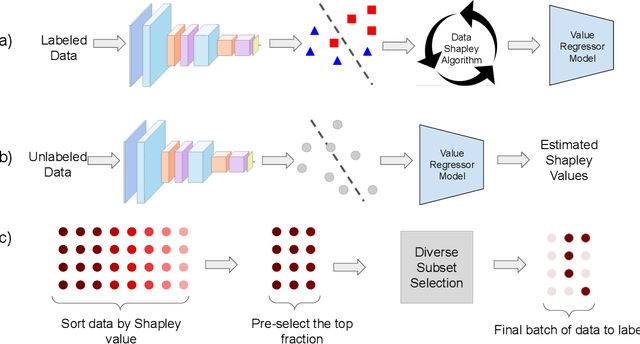
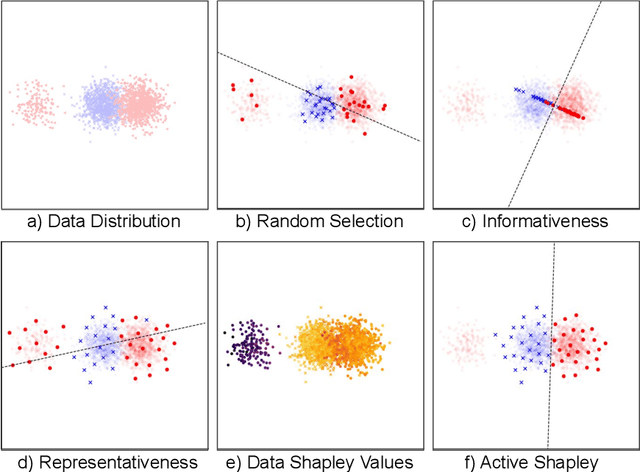
Abstract:Annotating the right set of data amongst all available data points is a key challenge in many machine learning applications. Batch active learning is a popular approach to address this, in which batches of unlabeled data points are selected for annotation, while an underlying learning algorithm gets subsequently updated. Increasingly larger batches are particularly appealing in settings where data can be annotated in parallel, and model training is computationally expensive. A key challenge here is scale - typical active learning methods rely on diversity techniques, which select a diverse set of data points to annotate, from an unlabeled pool. In this work, we introduce Active Data Shapley (ADS) -- a filtering layer for batch active learning that significantly increases the efficiency of active learning by pre-selecting, using a linear time computation, the highest-value points from an unlabeled dataset. Using the notion of the Shapley value of data, our method estimates the value of unlabeled data points with regards to the prediction task at hand. We show that ADS is particularly effective when the pool of unlabeled data exhibits real-world caveats: noise, heterogeneity, and domain shift. We run experiments demonstrating that when ADS is used to pre-select the highest-ranking portion of an unlabeled dataset, the efficiency of state-of-the-art batch active learning methods increases by an average factor of 6x, while preserving performance effectiveness.
Accurate Prediction of Free Solvation Energy of Organic Molecules via Graph Attention Network and Message Passing Neural Network from Pairwise Atomistic Interactions
Apr 15, 2021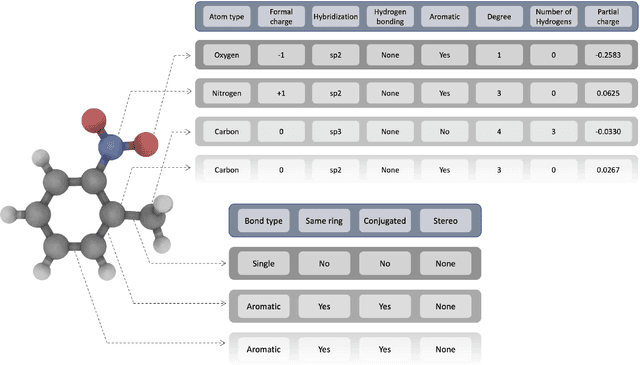
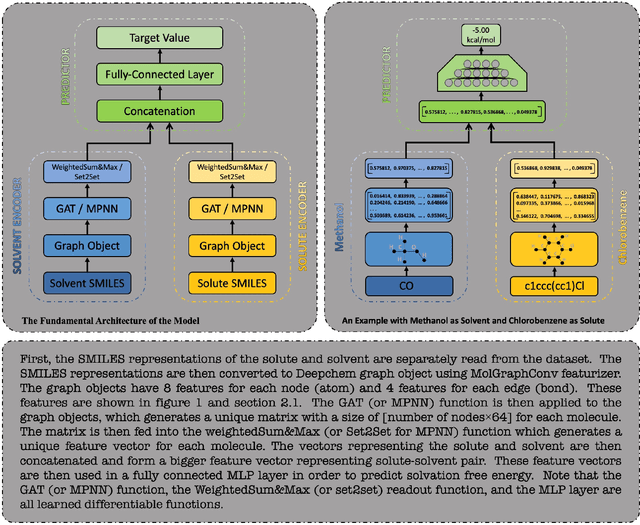

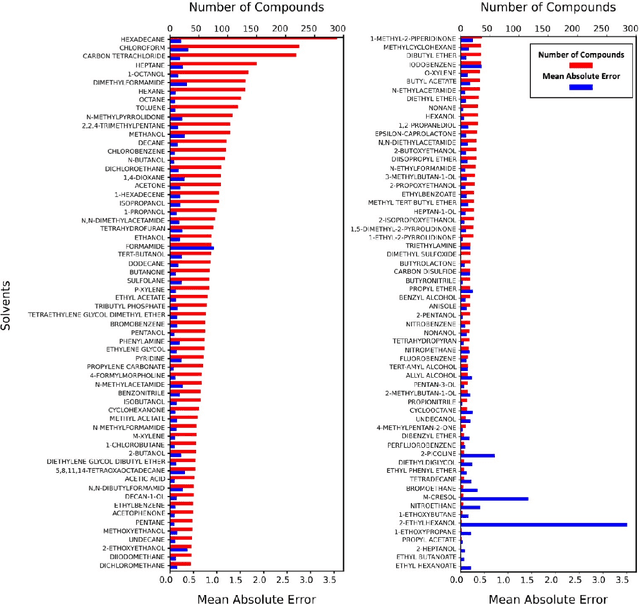
Abstract:Deep learning based methods have been widely applied to predict various kinds of molecular properties in the pharmaceutical industry with increasingly more success. Solvation free energy is an important index in the field of organic synthesis, medicinal chemistry, drug delivery, and biological processes. However, accurate solvation free energy determination is a time-consuming experimental process. Furthermore, it could be useful to assess solvation free energy in the absence of a physical sample. In this study, we propose two novel models for the problem of free solvation energy predictions, based on the Graph Neural Network (GNN) architectures: Message Passing Neural Network (MPNN) and Graph Attention Network (GAT). GNNs are capable of summarizing the predictive information of a molecule as low-dimensional features directly from its graph structure without relying on an extensive amount of intra-molecular descriptors. As a result, these models are capable of making accurate predictions of the molecular properties without the time consuming process of running an experiment on each molecule. We show that our proposed models outperform all quantum mechanical and molecular dynamics methods in addition to existing alternative machine learning based approaches in the task of solvation free energy prediction. We believe such promising predictive models will be applicable to enhancing the efficiency of the screening of drug molecules and be a useful tool to promote the development of molecular pharmaceutics.
Data Valuation for Medical Imaging Using Shapley Value: Application on A Large-scale Chest X-ray Dataset
Oct 15, 2020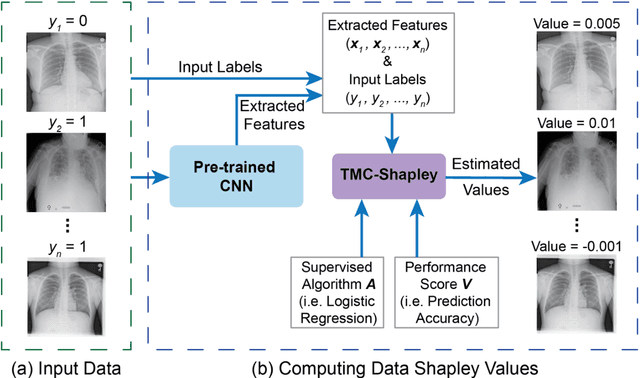
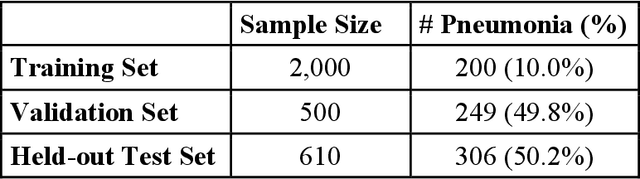
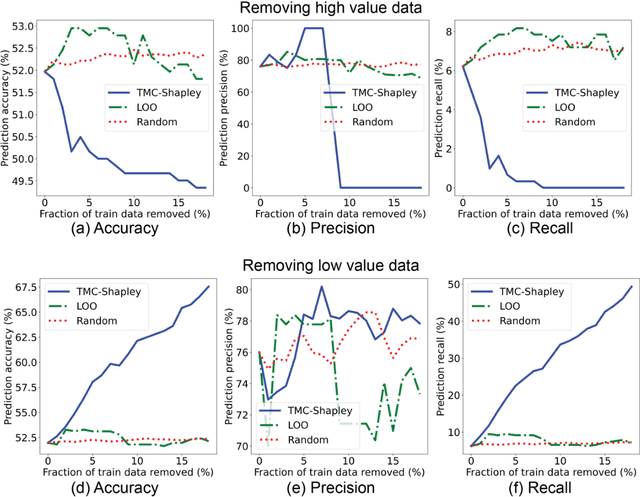

Abstract:The reliability of machine learning models can be compromised when trained on low quality data. Many large-scale medical imaging datasets contain low quality labels extracted from sources such as medical reports. Moreover, images within a dataset may have heterogeneous quality due to artifacts and biases arising from equipment or measurement errors. Therefore, algorithms that can automatically identify low quality data are highly desired. In this study, we used data Shapley, a data valuation metric, to quantify the value of training data to the performance of a pneumonia detection algorithm in a large chest X-ray dataset. We characterized the effectiveness of data Shapley in identifying low quality versus valuable data for pneumonia detection. We found that removing training data with high Shapley values decreased the pneumonia detection performance, whereas removing data with low Shapley values improved the model performance. Furthermore, there were more mislabeled examples in low Shapley value data and more true pneumonia cases in high Shapley value data. Our results suggest that low Shapley value indicates mislabeled or poor quality images, whereas high Shapley value indicates data that are valuable for pneumonia detection. Our method can serve as a framework for using data Shapley to denoise large-scale medical imaging datasets.
How Does Mixup Help With Robustness and Generalization?
Oct 09, 2020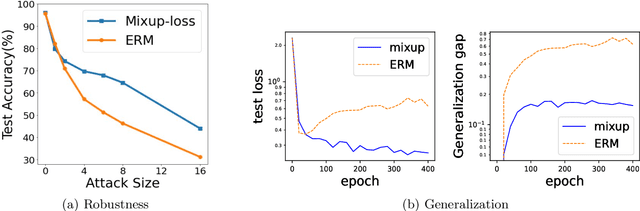
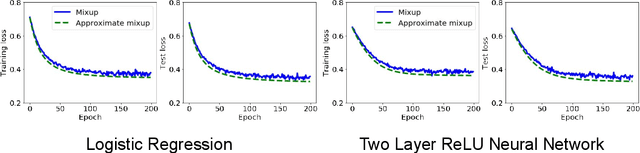
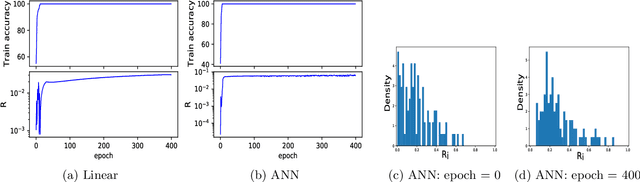
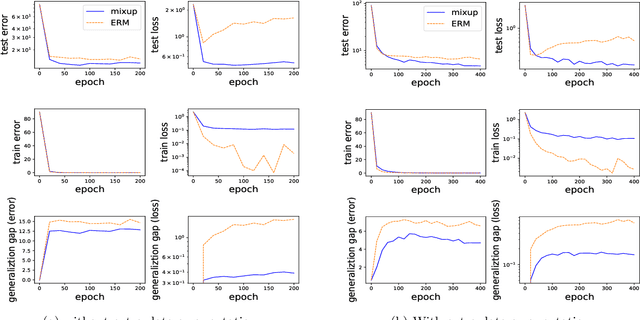
Abstract:Mixup is a popular data augmentation technique based on taking convex combinations of pairs of examples and their labels. This simple technique has been shown to substantially improve both the robustness and the generalization of the trained model. However, it is not well-understood why such improvement occurs. In this paper, we provide theoretical analysis to demonstrate how using Mixup in training helps model robustness and generalization. For robustness, we show that minimizing the Mixup loss corresponds to approximately minimizing an upper bound of the adversarial loss. This explains why models obtained by Mixup training exhibits robustness to several kinds of adversarial attacks such as Fast Gradient Sign Method (FGSM). For generalization, we prove that Mixup augmentation corresponds to a specific type of data-adaptive regularization which reduces overfitting. Our analysis provides new insights and a framework to understand Mixup.
Improving Adversarial Robustness via Unlabeled Out-of-Domain Data
Jun 15, 2020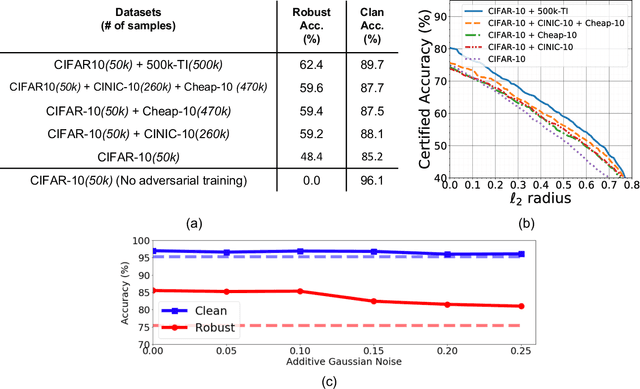

Abstract:Data augmentation by incorporating cheap unlabeled data from multiple domains is a powerful way to improve prediction especially when there is limited labeled data. In this work, we investigate how adversarial robustness can be enhanced by leveraging out-of-domain unlabeled data. We demonstrate that for broad classes of distributions and classifiers, there exists a sample complexity gap between standard and robust classification. We quantify to what degree this gap can be bridged via leveraging unlabeled samples from a shifted domain by providing both upper and lower bounds. Moreover, we show settings where we achieve better adversarial robustness when the unlabeled data come from a shifted domain rather than the same domain as the labeled data. We also investigate how to leverage out-of-domain data when some structural information, such as sparsity, is shared between labeled and unlabeled domains. Experimentally, we augment two object recognition datasets (CIFAR-10 and SVHN) with easy to obtain and unlabeled out-of-domain data and demonstrate substantial improvement in the model's robustness against $\ell_\infty$ adversarial attacks on the original domain.
A Distributional Framework for Data Valuation
Feb 27, 2020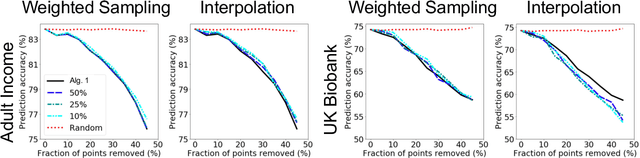
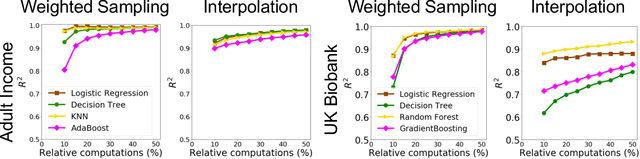
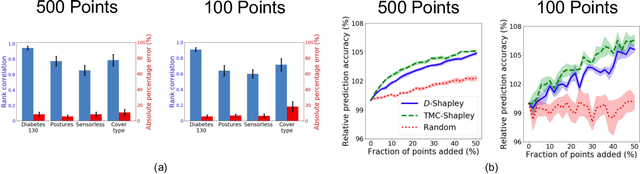
Abstract:Shapley value is a classic notion from game theory, historically used to quantify the contributions of individuals within groups, and more recently applied to assign values to data points when training machine learning models. Despite its foundational role, a key limitation of the data Shapley framework is that it only provides valuations for points within a fixed data set. It does not account for statistical aspects of the data and does not give a way to reason about points outside the data set. To address these limitations, we propose a novel framework -- distributional Shapley -- where the value of a point is defined in the context of an underlying data distribution. We prove that distributional Shapley has several desirable statistical properties; for example, the values are stable under perturbations to the data points themselves and to the underlying data distribution. We leverage these properties to develop a new algorithm for estimating values from data, which comes with formal guarantees and runs two orders of magnitude faster than state-of-the-art algorithms for computing the (non-distributional) data Shapley values. We apply distributional Shapley to diverse data sets and demonstrate its utility in a data market setting.
Neuron Shapley: Discovering the Responsible Neurons
Feb 25, 2020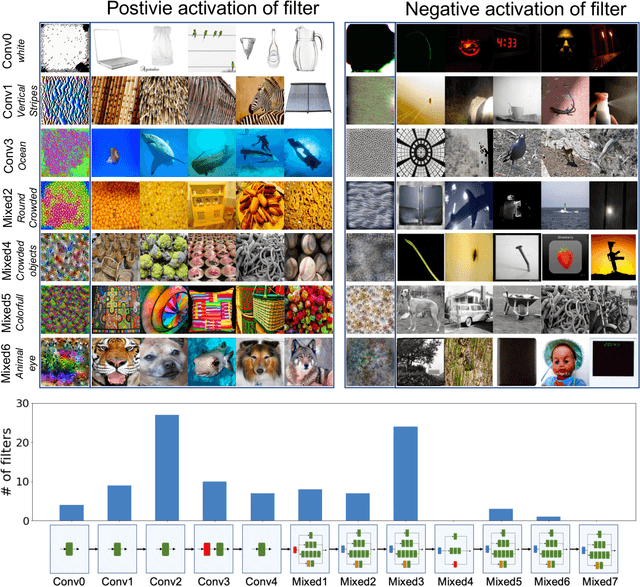
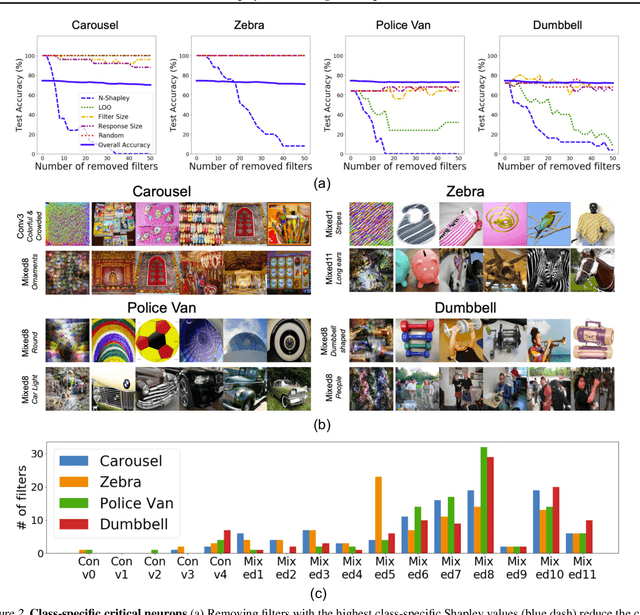
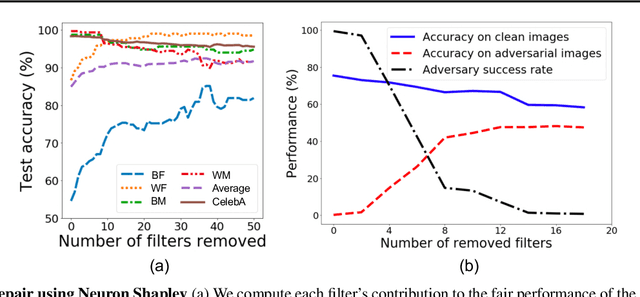
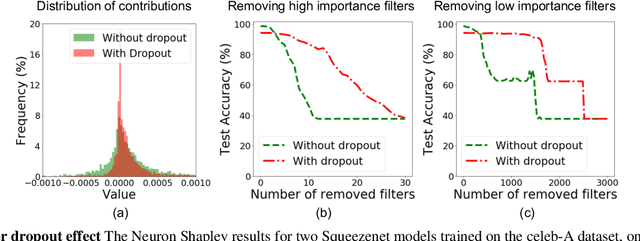
Abstract:We develop Neuron Shapley as a new framework to quantify the contribution of individual neurons to the prediction and performance of a deep network. By accounting for interactions across neurons, Neuron Shapley is more effective in identifying important filters compared to common approaches based on activation patterns. Interestingly, removing just 30 filters with the highest Shapley scores effectively destroys the prediction accuracy of Inception-v3 on ImageNet. Visualization of these few critical filters provides insights into how the network functions. Neuron Shapley is a flexible framework and can be applied to identify responsible neurons in many tasks. We illustrate additional applications of identifying filters that are responsible for biased prediction in facial recognition and filters that are vulnerable to adversarial attacks. Removing these filters is a quick way to repair models. Enabling all these applications is a new multi-arm bandit algorithm that we developed to efficiently estimate Neuron Shapley values.
DermGAN: Synthetic Generation of Clinical Skin Images with Pathology
Nov 20, 2019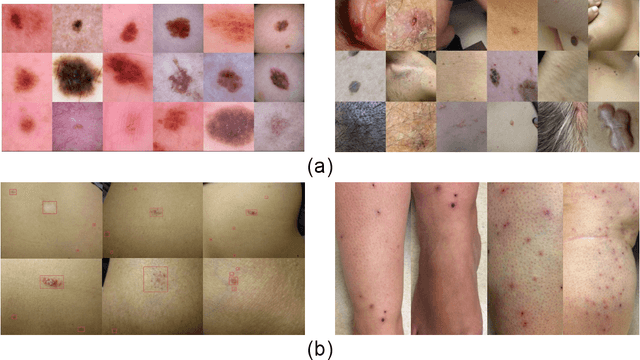

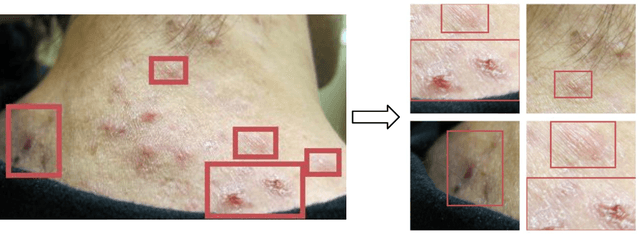
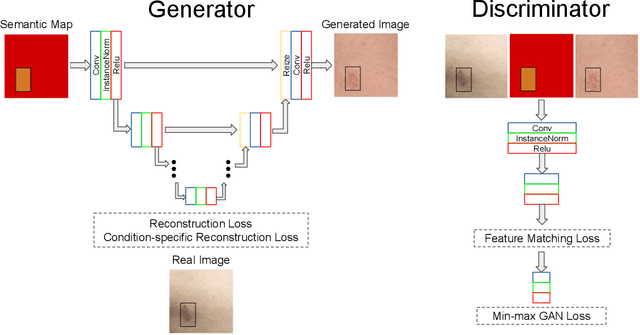
Abstract:Despite the recent success in applying supervised deep learning to medical imaging tasks, the problem of obtaining large and diverse expert-annotated datasets required for the development of high performant models remains particularly challenging. In this work, we explore the possibility of using Generative Adverserial Networks (GAN) to synthesize clinical images with skin condition. We propose DermGAN, an adaptation of the popular Pix2Pix architecture, to create synthetic images for a pre-specified skin condition while being able to vary its size, location and the underlying skin color. We demonstrate that the generated images are of high fidelity using objective GAN evaluation metrics. In a Human Turing test, we note that the synthetic images are not only visually similar to real images, but also embody the respective skin condition in dermatologists' eyes. Finally, when using the synthetic images as a data augmentation technique for training a skin condition classifier, we observe that the model performs comparably to the baseline model overall while improving on rare but malignant conditions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge