Andre Esteva
Improved Multimodal Fusion for Small Datasets with Auxiliary Supervision
Apr 01, 2023Abstract:Prostate cancer is one of the leading causes of cancer-related death in men worldwide. Like many cancers, diagnosis involves expert integration of heterogeneous patient information such as imaging, clinical risk factors, and more. For this reason, there have been many recent efforts toward deep multimodal fusion of image and non-image data for clinical decision tasks. Many of these studies propose methods to fuse learned features from each patient modality, providing significant downstream improvements with techniques like cross-modal attention gating, Kronecker product fusion, orthogonality regularization, and more. While these enhanced fusion operations can improve upon feature concatenation, they often come with an extremely high learning capacity, meaning they are likely to overfit when applied even to small or low-dimensional datasets. Rather than designing a highly expressive fusion operation, we propose three simple methods for improved multimodal fusion with small datasets that aid optimization by generating auxiliary sources of supervision during training: extra supervision, clinical prediction, and dense fusion. We validate the proposed approaches on prostate cancer diagnosis from paired histopathology imaging and tabular clinical features. The proposed methods are straightforward to implement and can be applied to any classification task with paired image and non-image data.
MetaHistoSeg: A Python Framework for Meta Learning in Histopathology Image Segmentation
Sep 29, 2021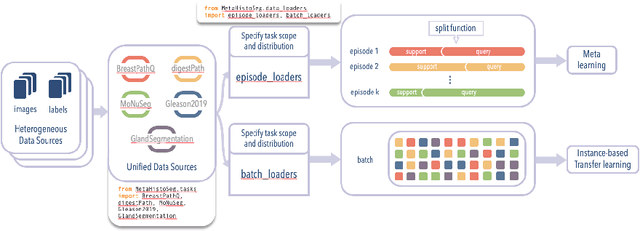
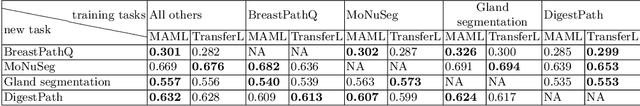
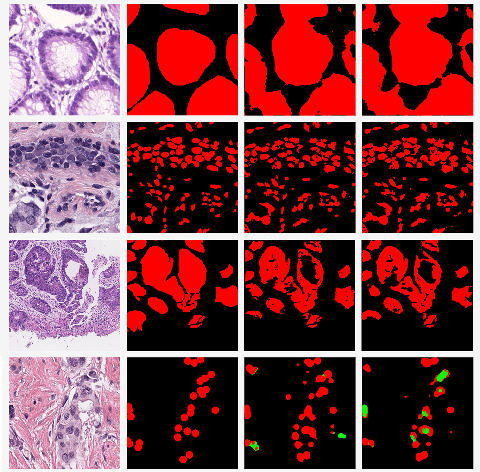
Abstract:Few-shot learning is a standard practice in most deep learning based histopathology image segmentation, given the relatively low number of digitized slides that are generally available. While many models have been developed for domain specific histopathology image segmentation, cross-domain generalization remains a key challenge for properly validating models. Here, tooling and datasets to benchmark model performance across histopathological domains are lacking. To address this limitation, we introduce MetaHistoSeg - a Python framework that implements unique scenarios in both meta learning and instance based transfer learning. Designed for easy extension to customized datasets and task sampling schemes, the framework empowers researchers with the ability of rapid model design and experimentation. We also curate a histopathology meta dataset - a benchmark dataset for training and validating models on out-of-distribution performance across a range of cancer types. In experiments we showcase the usage of MetaHistoSeg with the meta dataset and find that both meta-learning and instance based transfer learning deliver comparable results on average, but in some cases tasks can greatly benefit from one over the other.
Data Shapley Valuation for Efficient Batch Active Learning
Apr 16, 2021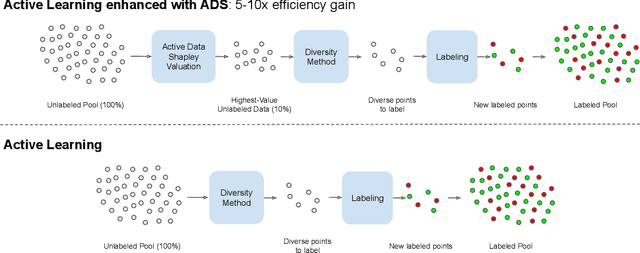
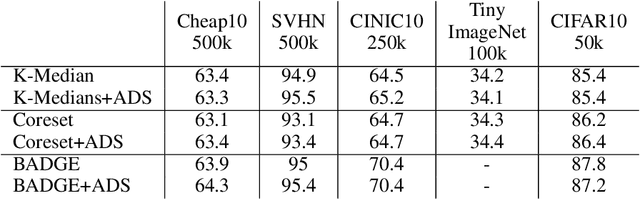
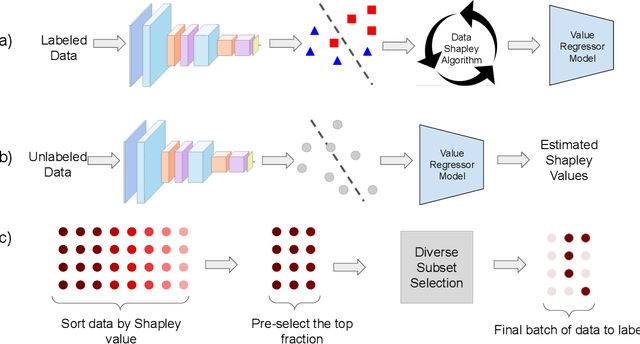
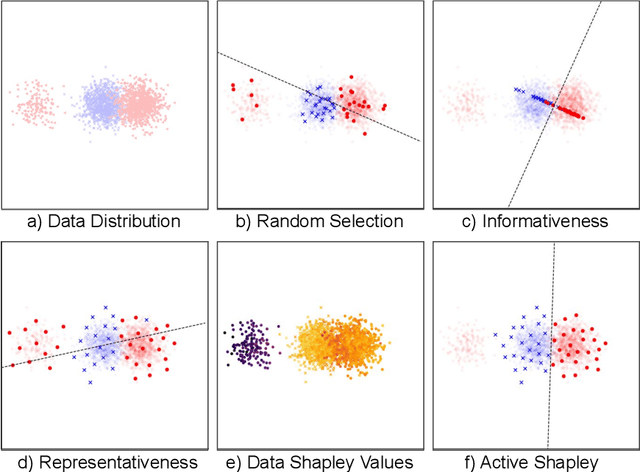
Abstract:Annotating the right set of data amongst all available data points is a key challenge in many machine learning applications. Batch active learning is a popular approach to address this, in which batches of unlabeled data points are selected for annotation, while an underlying learning algorithm gets subsequently updated. Increasingly larger batches are particularly appealing in settings where data can be annotated in parallel, and model training is computationally expensive. A key challenge here is scale - typical active learning methods rely on diversity techniques, which select a diverse set of data points to annotate, from an unlabeled pool. In this work, we introduce Active Data Shapley (ADS) -- a filtering layer for batch active learning that significantly increases the efficiency of active learning by pre-selecting, using a linear time computation, the highest-value points from an unlabeled dataset. Using the notion of the Shapley value of data, our method estimates the value of unlabeled data points with regards to the prediction task at hand. We show that ADS is particularly effective when the pool of unlabeled data exhibits real-world caveats: noise, heterogeneity, and domain shift. We run experiments demonstrating that when ADS is used to pre-select the highest-ranking portion of an unlabeled dataset, the efficiency of state-of-the-art batch active learning methods increases by an average factor of 6x, while preserving performance effectiveness.
CO-Search: COVID-19 Information Retrieval with Semantic Search, Question Answering, and Abstractive Summarization
Jun 17, 2020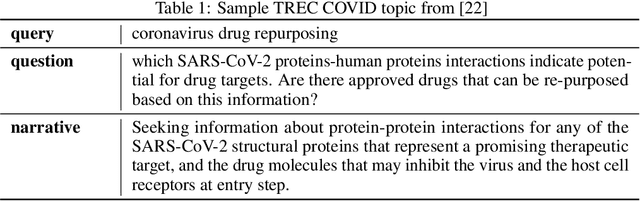
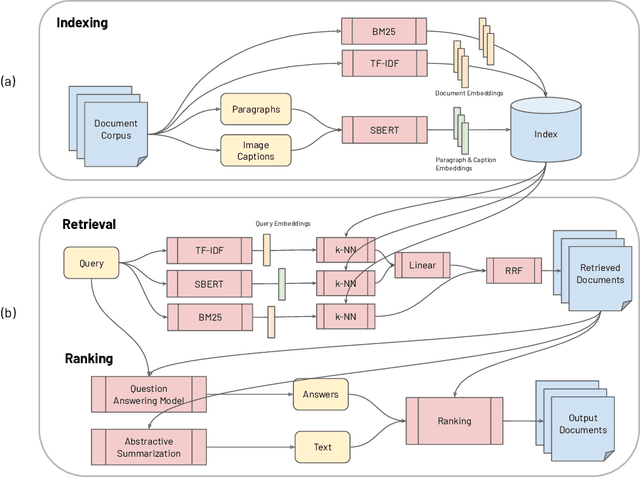
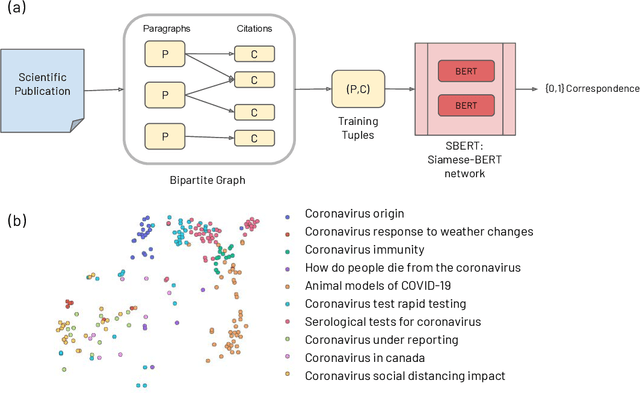

Abstract:The COVID-19 global pandemic has resulted in international efforts to understand, track, and mitigate the disease, yielding a significant corpus of COVID-19 and SARS-CoV-2-related publications across scientific disciplines. As of May 2020, 128,000 coronavirus-related publications have been collected through the COVID-19 Open Research Dataset Challenge. Here we present CO-Search, a retriever-ranker semantic search engine designed to handle complex queries over the COVID-19 literature, potentially aiding overburdened health workers in finding scientific answers during a time of crisis. The retriever is built from a Siamese-BERT encoder that is linearly composed with a TF-IDF vectorizer, and reciprocal-rank fused with a BM25 vectorizer. The ranker is composed of a multi-hop question-answering module, that together with a multi-paragraph abstractive summarizer adjust retriever scores. To account for the domain-specific and relatively limited dataset, we generate a bipartite graph of document paragraphs and citations, creating 1.3 million (citation title, paragraph) tuples for training the encoder. We evaluate our system on the data of the TREC-COVID information retrieval challenge. CO-Search obtains top performance on the datasets of the first and second rounds, across several key metrics: normalized discounted cumulative gain, precision, mean average precision, and binary preference.
Skin Cancer Detection and Tracking using Data Synthesis and Deep Learning
Dec 04, 2016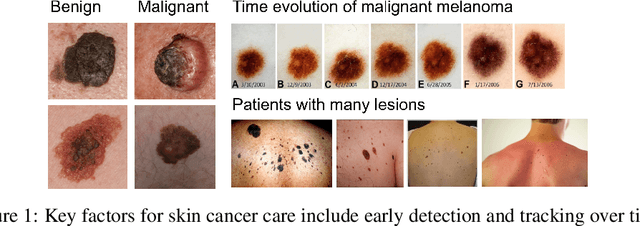
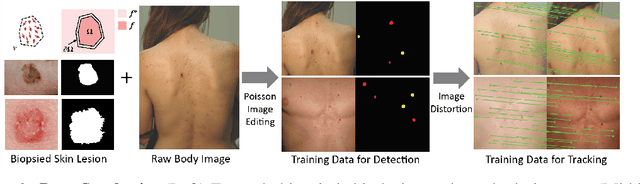
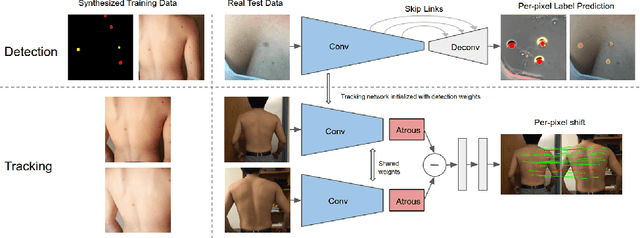
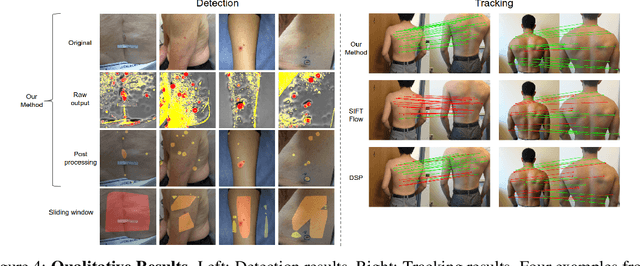
Abstract:Dense object detection and temporal tracking are needed across applications domains ranging from people-tracking to analysis of satellite imagery over time. The detection and tracking of malignant skin cancers and benign moles poses a particularly challenging problem due to the general uniformity of large skin patches, the fact that skin lesions vary little in their appearance, and the relatively small amount of data available. Here we introduce a novel data synthesis technique that merges images of individual skin lesions with full-body images and heavily augments them to generate significant amounts of data. We build a convolutional neural network (CNN) based system, trained on this synthetic data, and demonstrate superior performance to traditional detection and tracking techniques. Additionally, we compare our system to humans trained with simple criteria. Our system is intended for potential clinical use to augment the capabilities of healthcare providers. While domain-specific, we believe the methods invoked in this work will be useful in applying CNNs across domains that suffer from limited data availability.
Affordances Provide a Fundamental Categorization Principle for Visual Scenes
Nov 19, 2014Abstract:How do we know that a kitchen is a kitchen by looking? Relatively little is known about how we conceptualize and categorize different visual environments. Traditional models of visual perception posit that scene categorization is achieved through the recognition of a scene's objects, yet these models cannot account for the mounting evidence that human observers are relatively insensitive to the local details in an image. Psychologists have long theorized that the affordances, or actionable possibilities of a stimulus are pivotal to its perception. To what extent are scene categories created from similar affordances? Using a large-scale experiment using hundreds of scene categories, we show that the activities afforded by a visual scene provide a fundamental categorization principle. Affordance-based similarity explained the majority of the structure in the human scene categorization patterns, outperforming alternative similarities based on objects or visual features. We all models were combined, affordances provided the majority of the predictive power in the combined model, and nearly half of the total explained variance is captured only by affordances. These results challenge many existing models of high-level visual perception, and provide immediately testable hypotheses for the functional organization of the human perceptual system.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge