Sebastian Thrun
Restructuring Vector Quantization with the Rotation Trick
Oct 08, 2024



Abstract:Vector Quantized Variational AutoEncoders (VQ-VAEs) are designed to compress a continuous input to a discrete latent space and reconstruct it with minimal distortion. They operate by maintaining a set of vectors -- often referred to as the codebook -- and quantizing each encoder output to the nearest vector in the codebook. However, as vector quantization is non-differentiable, the gradient to the encoder flows around the vector quantization layer rather than through it in a straight-through approximation. This approximation may be undesirable as all information from the vector quantization operation is lost. In this work, we propose a way to propagate gradients through the vector quantization layer of VQ-VAEs. We smoothly transform each encoder output into its corresponding codebook vector via a rotation and rescaling linear transformation that is treated as a constant during backpropagation. As a result, the relative magnitude and angle between encoder output and codebook vector becomes encoded into the gradient as it propagates through the vector quantization layer and back to the encoder. Across 11 different VQ-VAE training paradigms, we find this restructuring improves reconstruction metrics, codebook utilization, and quantization error. Our code is available at https://github.com/cfifty/rotation_trick.
BanditPAM++: Faster $k$-medoids Clustering
Oct 28, 2023


Abstract:Clustering is a fundamental task in data science with wide-ranging applications. In $k$-medoids clustering, cluster centers must be actual datapoints and arbitrary distance metrics may be used; these features allow for greater interpretability of the cluster centers and the clustering of exotic objects in $k$-medoids clustering, respectively. $k$-medoids clustering has recently grown in popularity due to the discovery of more efficient $k$-medoids algorithms. In particular, recent research has proposed BanditPAM, a randomized $k$-medoids algorithm with state-of-the-art complexity and clustering accuracy. In this paper, we present BanditPAM++, which accelerates BanditPAM via two algorithmic improvements, and is $O(k)$ faster than BanditPAM in complexity and substantially faster than BanditPAM in wall-clock runtime. First, we demonstrate that BanditPAM has a special structure that allows the reuse of clustering information $\textit{within}$ each iteration. Second, we demonstrate that BanditPAM has additional structure that permits the reuse of information $\textit{across}$ different iterations. These observations inspire our proposed algorithm, BanditPAM++, which returns the same clustering solutions as BanditPAM but often several times faster. For example, on the CIFAR10 dataset, BanditPAM++ returns the same results as BanditPAM but runs over 10$\times$ faster. Finally, we provide a high-performance C++ implementation of BanditPAM++, callable from Python and R, that may be of interest to practitioners at https://github.com/motiwari/BanditPAM. Auxiliary code to reproduce all of our experiments via a one-line script is available at https://github.com/ThrunGroup/BanditPAM_plusplus_experiments.
Context-Aware Meta-Learning
Oct 17, 2023



Abstract:Large Language Models like ChatGPT demonstrate a remarkable capacity to learn new concepts during inference without any fine-tuning. However, visual models trained to detect new objects during inference have been unable to replicate this ability, and instead either perform poorly or require meta-training and/or fine-tuning on similar objects. In this work, we propose a meta-learning algorithm that emulates Large Language Models by learning new visual concepts during inference without fine-tuning. Our approach leverages a frozen pre-trained feature extractor, and analogous to in-context learning, recasts meta-learning as sequence modeling over datapoints with known labels and a test datapoint with an unknown label. On 8 out of 11 meta-learning benchmarks, our approach -- without meta-training or fine-tuning -- exceeds or matches the state-of-the-art algorithm, P>M>F, which is meta-trained on these benchmarks.
In-Context Learning for Few-Shot Molecular Property Prediction
Oct 13, 2023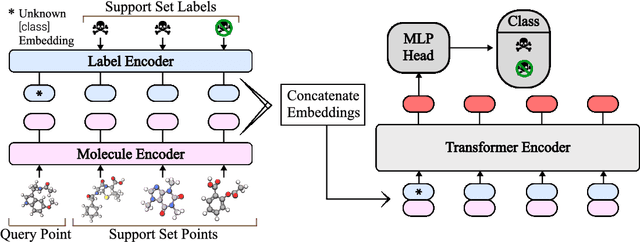
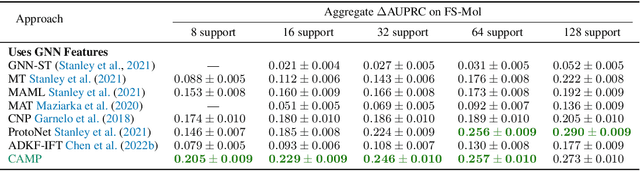
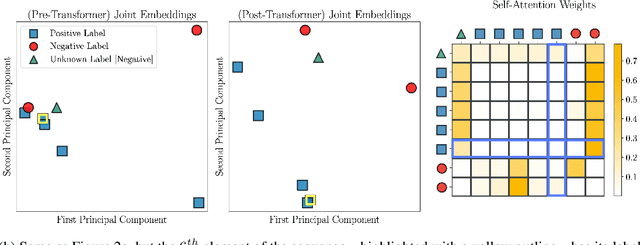
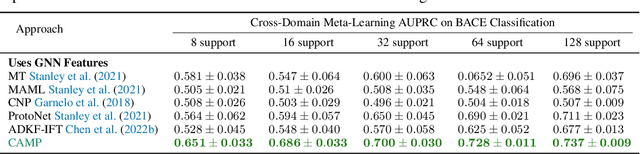
Abstract:In-context learning has become an important approach for few-shot learning in Large Language Models because of its ability to rapidly adapt to new tasks without fine-tuning model parameters. However, it is restricted to applications in natural language and inapplicable to other domains. In this paper, we adapt the concepts underpinning in-context learning to develop a new algorithm for few-shot molecular property prediction. Our approach learns to predict molecular properties from a context of (molecule, property measurement) pairs and rapidly adapts to new properties without fine-tuning. On the FS-Mol and BACE molecular property prediction benchmarks, we find this method surpasses the performance of recent meta-learning algorithms at small support sizes and is competitive with the best methods at large support sizes.
MAPTree: Beating "Optimal" Decision Trees with Bayesian Decision Trees
Sep 26, 2023



Abstract:Decision trees remain one of the most popular machine learning models today, largely due to their out-of-the-box performance and interpretability. In this work, we present a Bayesian approach to decision tree induction via maximum a posteriori inference of a posterior distribution over trees. We first demonstrate a connection between maximum a posteriori inference of decision trees and AND/OR search. Using this connection, we propose an AND/OR search algorithm, dubbed MAPTree, which is able to recover the maximum a posteriori tree. Lastly, we demonstrate the empirical performance of the maximum a posteriori tree both on synthetic data and in real world settings. On 16 real world datasets, MAPTree either outperforms baselines or demonstrates comparable performance but with much smaller trees. On a synthetic dataset, MAPTree also demonstrates greater robustness to noise and better generalization than existing approaches. Finally, MAPTree recovers the maxiumum a posteriori tree faster than existing sampling approaches and, in contrast with those algorithms, is able to provide a certificate of optimality. The code for our experiments is available at https://github.com/ThrunGroup/maptree.
Bayesian Decision Trees via Tractable Priors and Probabilistic Context-Free Grammars
Feb 15, 2023Abstract:Decision Trees are some of the most popular machine learning models today due to their out-of-the-box performance and interpretability. Often, Decision Trees models are constructed greedily in a top-down fashion via heuristic search criteria, such as Gini impurity or entropy. However, trees constructed in this manner are sensitive to minor fluctuations in training data and are prone to overfitting. In contrast, Bayesian approaches to tree construction formulate the selection process as a posterior inference problem; such approaches are more stable and provide greater theoretical guarantees. However, generating Bayesian Decision Trees usually requires sampling from complex, multimodal posterior distributions. Current Markov Chain Monte Carlo-based approaches for sampling Bayesian Decision Trees are prone to mode collapse and long mixing times, which makes them impractical. In this paper, we propose a new criterion for training Bayesian Decision Trees. Our criterion gives rise to BCART-PCFG, which can efficiently sample decision trees from a posterior distribution across trees given the data and find the maximum a posteriori (MAP) tree. Learning the posterior and training the sampler can be done in time that is polynomial in the dataset size. Once the posterior has been learned, trees can be sampled efficiently (linearly in the number of nodes). At the core of our method is a reduction of sampling the posterior to sampling a derivation from a probabilistic context-free grammar. We find that trees sampled via BCART-PCFG perform comparable to or better than greedily-constructed Decision Trees in classification accuracy on several datasets. Additionally, the trees sampled via BCART-PCFG are significantly smaller -- sometimes by as much as 20x.
MABSplit: Faster Forest Training Using Multi-Armed Bandits
Dec 14, 2022



Abstract:Random forests are some of the most widely used machine learning models today, especially in domains that necessitate interpretability. We present an algorithm that accelerates the training of random forests and other popular tree-based learning methods. At the core of our algorithm is a novel node-splitting subroutine, dubbed MABSplit, used to efficiently find split points when constructing decision trees. Our algorithm borrows techniques from the multi-armed bandit literature to judiciously determine how to allocate samples and computational power across candidate split points. We provide theoretical guarantees that MABSplit improves the sample complexity of each node split from linear to logarithmic in the number of data points. In some settings, MABSplit leads to 100x faster training (an 99% reduction in training time) without any decrease in generalization performance. We demonstrate similar speedups when MABSplit is used across a variety of forest-based variants, such as Extremely Random Forests and Random Patches. We also show our algorithm can be used in both classification and regression tasks. Finally, we show that MABSplit outperforms existing methods in generalization performance and feature importance calculations under a fixed computational budget. All of our experimental results are reproducible via a one-line script at https://github.com/ThrunGroup/FastForest.
Faster Maximum Inner Product Search in High Dimensions
Dec 14, 2022Abstract:Maximum Inner Product Search (MIPS) is a popular problem in the machine learning literature due to its applicability in a wide array of applications, such as recommender systems. In high-dimensional settings, however, MIPS queries can become computationally expensive as most existing solutions do not scale well with data dimensionality. In this work, we present a state-of-the-art algorithm for the MIPS problem in high dimensions, dubbed BanditMIPS. BanditMIPS is a randomized algorithm that borrows techniques from multi-armed bandits to reduce the MIPS problem to a best-arm identification problem. BanditMIPS reduces the complexity of state-of-the-art algorithms from $O(\sqrt{d})$ to $O(\text{log}d)$, where $d$ is the dimension of the problem data vectors. On high-dimensional real-world datasets, BanditMIPS runs approximately 12 times faster than existing approaches and returns the same solution. BanditMIPS requires no preprocessing of the data and includes a hyperparameter that practitioners may use to trade off accuracy and runtime. We also propose a variant of our algorithm, named BanditMIPS-$\alpha$, which employs non-uniform sampling across the data dimensions to provide further speedups.
Bandit-PAM: Almost Linear Time $k$-Medoids Clustering via Multi-Armed Bandits
Jun 11, 2020

Abstract:Clustering is a ubiquitous task in data science. Compared to the commonly used $k$-means clustering algorithm, $k$-medoids clustering algorithms require the cluster centers to be actual data points and support arbitrary distance metrics, allowing for greater interpretability and the clustering of structured objects. Current state-of-the-art $k$-medoids clustering algorithms, such as Partitioning Around Medoids (PAM), are iterative and are quadratic in the dataset size $n$ for each iteration, being prohibitively expensive for large datasets. We propose Bandit-PAM, a randomized algorithm inspired by techniques from multi-armed bandits, that significantly improves the computational efficiency of PAM. We theoretically prove that Bandit-PAM reduces the complexity of each PAM iteration from $O(n^2)$ to $O(n \log n)$ and returns the same results with high probability, under assumptions on the data that often hold in practice. We empirically validate our results on several large-scale real-world datasets, including a coding exercise submissions dataset from Code.org, the 10x Genomics 68k PBMC single-cell RNA sequencing dataset, and the MNIST handwritten digits dataset. We observe that Bandit-PAM returns the same results as PAM while performing up to 200x fewer distance computations. The improvements demonstrated by Bandit-PAM enable $k$-medoids clustering on a wide range of applications, including identifying cell types in large-scale single-cell data and providing scalable feedback for students learning computer science online. We also release Python and C++ implementations of our algorithm.
Skin Cancer Detection and Tracking using Data Synthesis and Deep Learning
Dec 04, 2016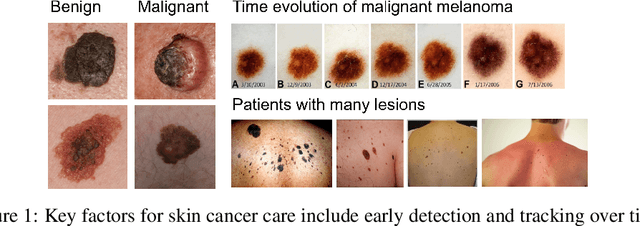
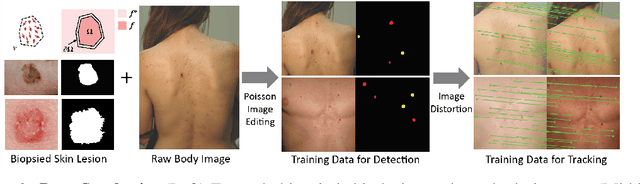
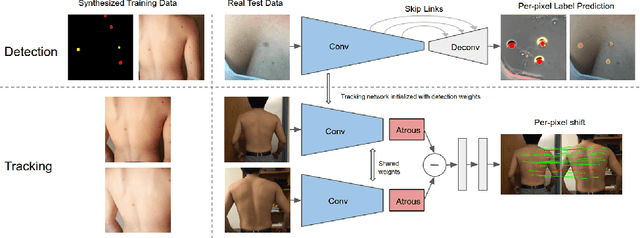
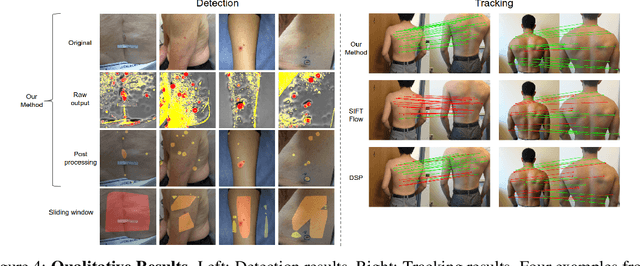
Abstract:Dense object detection and temporal tracking are needed across applications domains ranging from people-tracking to analysis of satellite imagery over time. The detection and tracking of malignant skin cancers and benign moles poses a particularly challenging problem due to the general uniformity of large skin patches, the fact that skin lesions vary little in their appearance, and the relatively small amount of data available. Here we introduce a novel data synthesis technique that merges images of individual skin lesions with full-body images and heavily augments them to generate significant amounts of data. We build a convolutional neural network (CNN) based system, trained on this synthetic data, and demonstrate superior performance to traditional detection and tracking techniques. Additionally, we compare our system to humans trained with simple criteria. Our system is intended for potential clinical use to augment the capabilities of healthcare providers. While domain-specific, we believe the methods invoked in this work will be useful in applying CNNs across domains that suffer from limited data availability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge