Jennifer I. Lim
Multi-OCT-SelfNet: Integrating Self-Supervised Learning with Multi-Source Data Fusion for Enhanced Multi-Class Retinal Disease Classification
Sep 17, 2024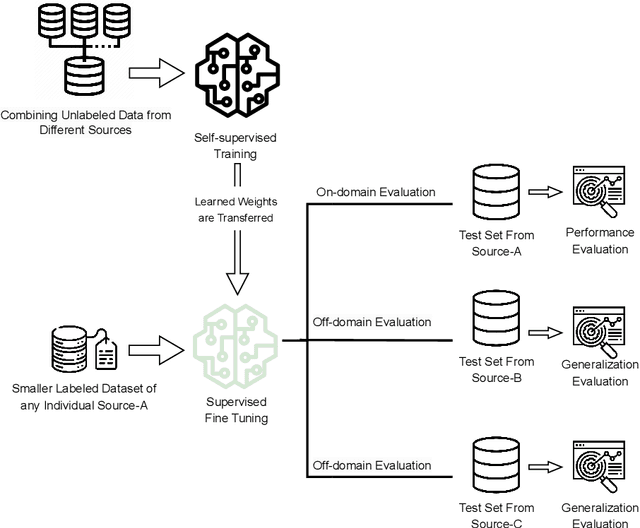
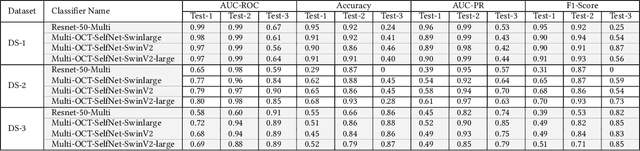
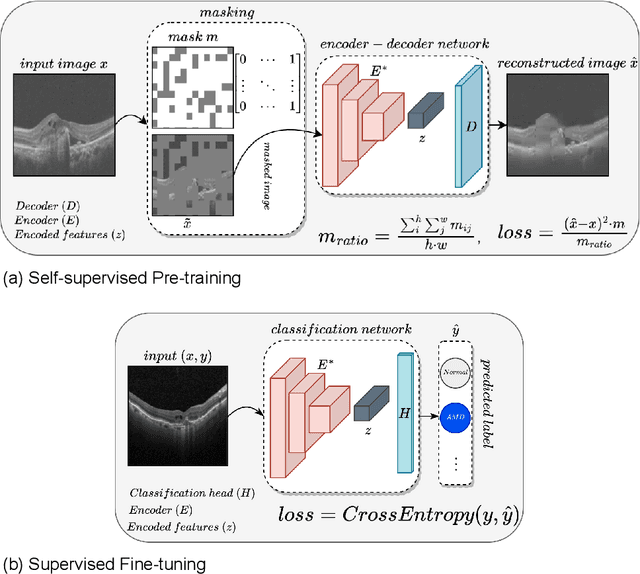
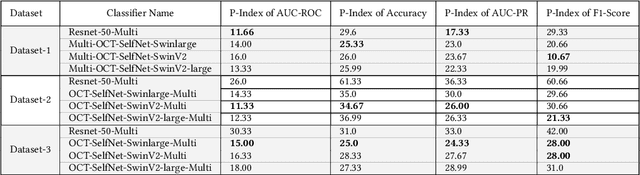
Abstract:In the medical domain, acquiring large datasets poses significant challenges due to privacy concerns. Nonetheless, the development of a robust deep-learning model for retinal disease diagnosis necessitates a substantial dataset for training. The capacity to generalize effectively on smaller datasets remains a persistent challenge. The scarcity of data presents a significant barrier to the practical implementation of scalable medical AI solutions. To address this issue, we've combined a wide range of data sources to improve performance and generalization to new data by giving it a deeper understanding of the data representation from multi-modal datasets and developed a self-supervised framework based on large language models (LLMs), SwinV2 to gain a deeper understanding of multi-modal dataset representations, enhancing the model's ability to extrapolate to new data for the detection of eye diseases using optical coherence tomography (OCT) images. We adopt a two-phase training methodology, self-supervised pre-training, and fine-tuning on a downstream supervised classifier. An ablation study conducted across three datasets employing various encoder backbones, without data fusion, with low data availability setting, and without self-supervised pre-training scenarios, highlights the robustness of our method. Our findings demonstrate consistent performance across these diverse conditions, showcasing superior generalization capabilities compared to the baseline model, ResNet-50.
Quantitative Characterization of Retinal Features in Translated OCTA
Apr 24, 2024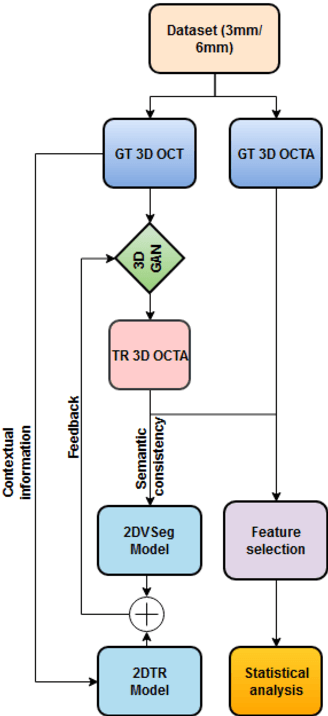
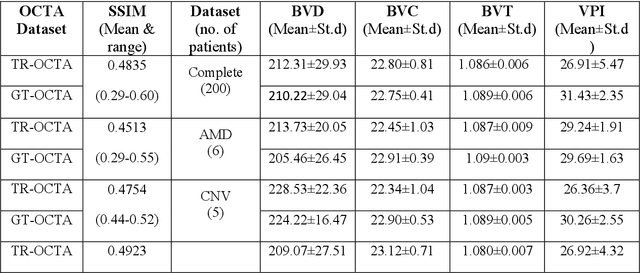
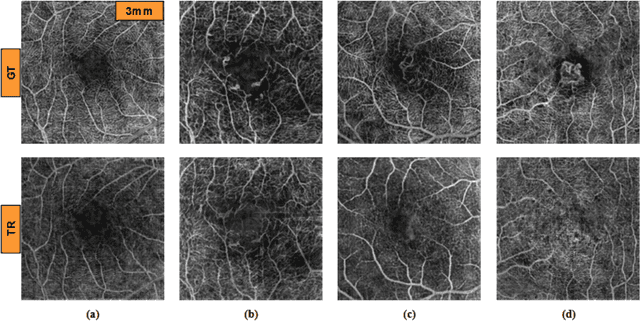
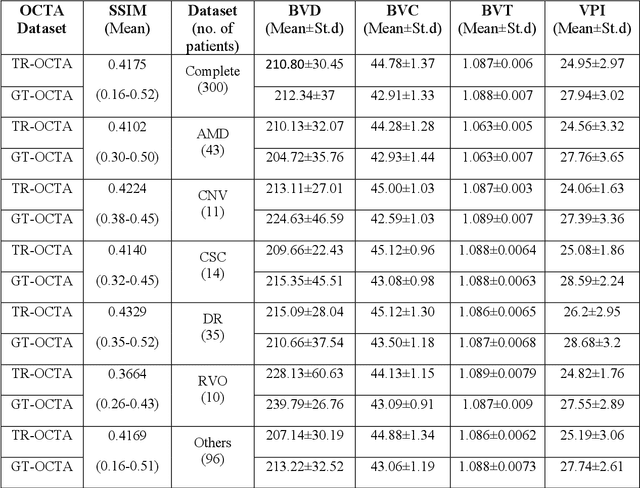
Abstract:Purpose: This study explores the feasibility of using generative machine learning (ML) to translate Optical Coherence Tomography (OCT) images into Optical Coherence Tomography Angiography (OCTA) images, potentially bypassing the need for specialized OCTA hardware. Methods: The method involved implementing a generative adversarial network framework that includes a 2D vascular segmentation model and a 2D OCTA image translation model. The study utilizes a public dataset of 500 patients, divided into subsets based on resolution and disease status, to validate the quality of TR-OCTA images. The validation employs several quality and quantitative metrics to compare the translated images with ground truth OCTAs (GT-OCTA). We then quantitatively characterize vascular features generated in TR-OCTAs with GT-OCTAs to assess the feasibility of using TR-OCTA for objective disease diagnosis. Result: TR-OCTAs showed high image quality in both 3 and 6 mm datasets (high-resolution, moderate structural similarity and contrast quality compared to GT-OCTAs). There were slight discrepancies in vascular metrics, especially in diseased patients. Blood vessel features like tortuosity and vessel perimeter index showed a better trend compared to density features which are affected by local vascular distortions. Conclusion: This study presents a promising solution to the limitations of OCTA adoption in clinical practice by using vascular features from TR-OCTA for disease detection. Translation relevance: This study has the potential to significantly enhance the diagnostic process for retinal diseases by making detailed vascular imaging more widely available and reducing dependency on costly OCTA equipment.
Normalized Blood Flow Index in Optical Coherence Tomography Angiography Provides a Sensitive Biomarker of Early Diabetic Retinopathy
Dec 22, 2022Abstract:Purpose: To evaluate the sensitivity of normalized blood flow index (NBFI) for detecting early diabetic retinopathy (DR). Methods: Optical coherence tomography angiography (OCTA) images of 30 eyes from 20 healthy controls, 21 eyes of diabetic patients with no DR (NoDR) and 26 eyes from 22 patients with mild non-proliferative DR (NPDR) were analyzed in this study. The OCTA images were centered on the fovea and covered a 6 mm x 6 mm area. Enface projections of the superficial vascular plexus (SVP) and the deep capillary plexus (DCP) were obtained for the quantitative OCTA feature analysis. Three quantitative OCTA features were examined: blood vessel density (BVD), blood flow flux (BFF), and normalized blood flow index (NBFI). Each feature was calculated from both the SVP and DCP and their sensitivity to distinguish the three cohorts of the study were evaluated. Results: The only quantitative feature that was capable of distinguishing between all three cohorts was NBFI in the DCP image. Comparative study revealed that both BVD and BFF were able to distinguish the controls from NoDR and mild NPDR. However, neither BVD nor BFF was sensitive enough to separate NoDR from the healthy controls. Conclusion: The NBFI has been demonstrated as a sensitive biomarker of early DR, revealing retinal blood flow abnormality better than traditional BVD and BFF. The NBFI in the DCP was verified as the most sensitive biomarker, supporting that diabetes affects the DCP earlier than SVP in DR.
Contrastive learning-based pretraining improves representation and transferability of diabetic retinopathy classification models
Aug 24, 2022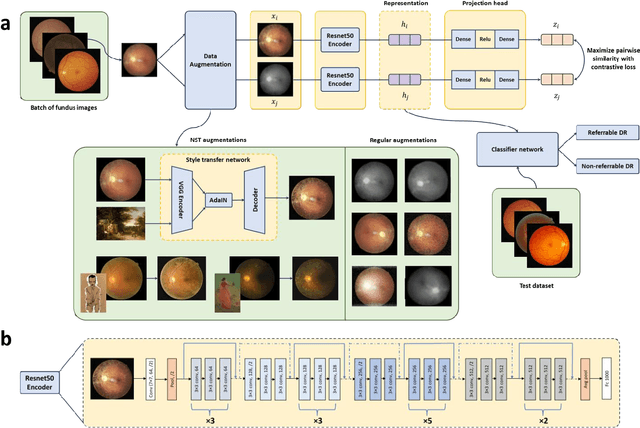
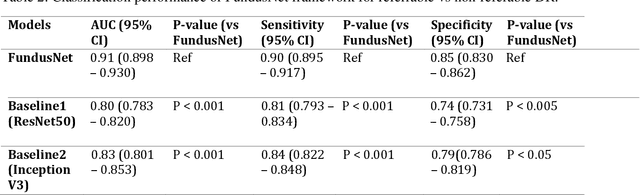
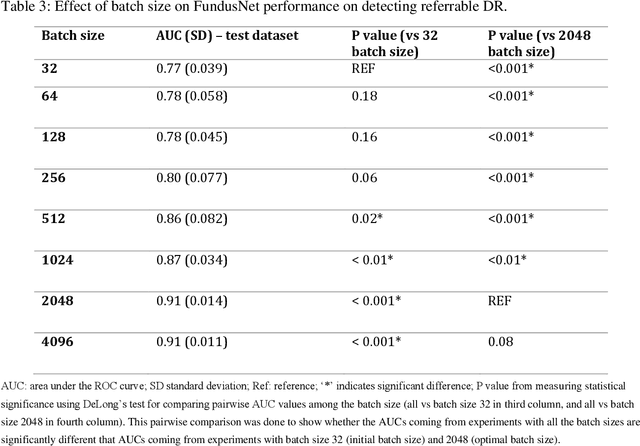
Abstract:Self supervised contrastive learning based pretraining allows development of robust and generalized deep learning models with small, labeled datasets, reducing the burden of label generation. This paper aims to evaluate the effect of CL based pretraining on the performance of referrable vs non referrable diabetic retinopathy (DR) classification. We have developed a CL based framework with neural style transfer (NST) augmentation to produce models with better representations and initializations for the detection of DR in color fundus images. We compare our CL pretrained model performance with two state of the art baseline models pretrained with Imagenet weights. We further investigate the model performance with reduced labeled training data (down to 10 percent) to test the robustness of the model when trained with small, labeled datasets. The model is trained and validated on the EyePACS dataset and tested independently on clinical data from the University of Illinois, Chicago (UIC). Compared to baseline models, our CL pretrained FundusNet model had higher AUC (CI) values (0.91 (0.898 to 0.930) vs 0.80 (0.783 to 0.820) and 0.83 (0.801 to 0.853) on UIC data). At 10 percent labeled training data, the FundusNet AUC was 0.81 (0.78 to 0.84) vs 0.58 (0.56 to 0.64) and 0.63 (0.60 to 0.66) in baseline models, when tested on the UIC dataset. CL based pretraining with NST significantly improves DL classification performance, helps the model generalize well (transferable from EyePACS to UIC data), and allows training with small, annotated datasets, therefore reducing ground truth annotation burden of the clinicians.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge