Jin Long
Critical Evaluation of Artificial Intelligence as Digital Twin of Pathologist for Prostate Cancer Pathology
Aug 23, 2023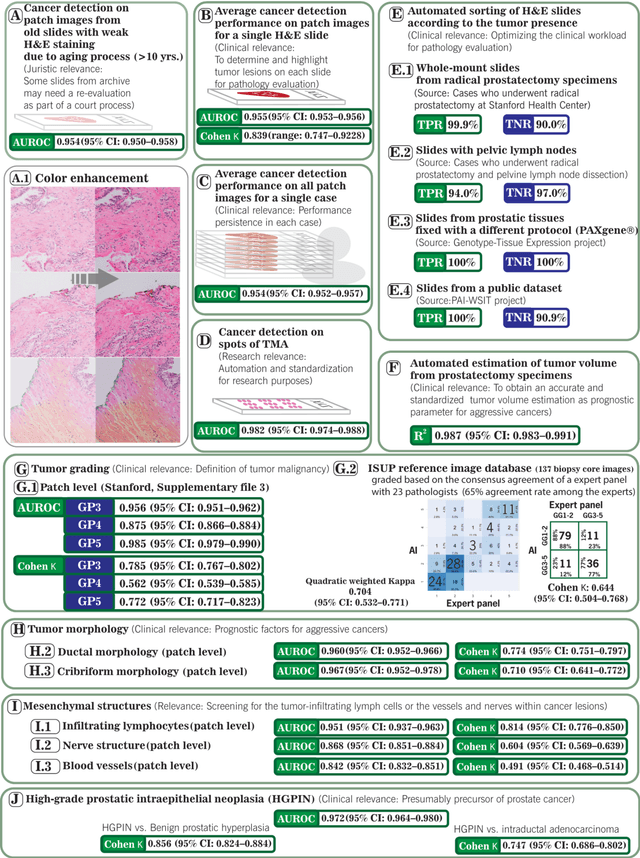
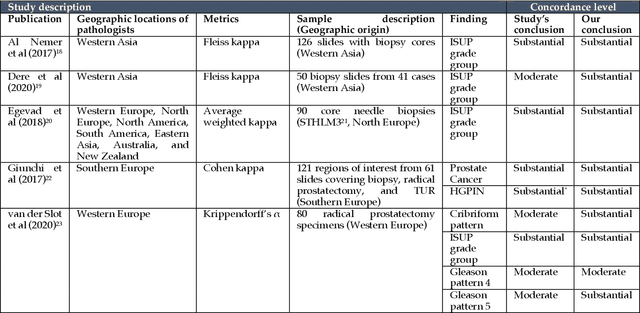
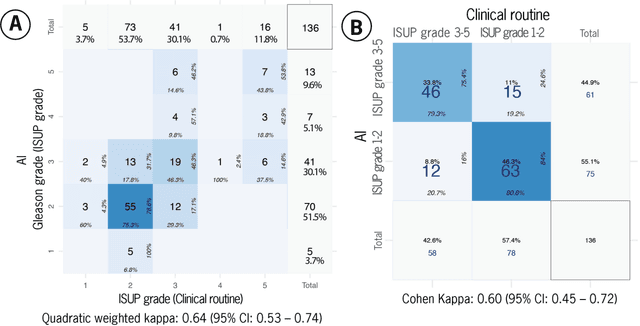
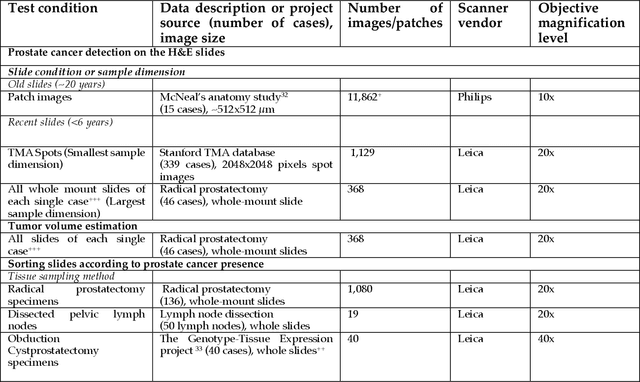
Abstract:Prostate cancer pathology plays a crucial role in clinical management but is time-consuming. Artificial intelligence (AI) shows promise in detecting prostate cancer and grading patterns. We tested an AI-based digital twin of a pathologist, vPatho, on 2,603 histology images of prostate tissue stained with hematoxylin and eosin. We analyzed various factors influencing tumor-grade disagreement between vPatho and six human pathologists. Our results demonstrated that vPatho achieved comparable performance in prostate cancer detection and tumor volume estimation, as reported in the literature. Concordance levels between vPatho and human pathologists were examined. Notably, moderate to substantial agreement was observed in identifying complementary histological features such as ductal, cribriform, nerve, blood vessels, and lymph cell infiltrations. However, concordance in tumor grading showed a decline when applied to prostatectomy specimens (kappa = 0.44) compared to biopsy cores (kappa = 0.70). Adjusting the decision threshold for the secondary Gleason pattern from 5% to 10% improved the concordance level between pathologists and vPatho for tumor grading on prostatectomy specimens (kappa from 0.44 to 0.64). Potential causes of grade discordance included the vertical extent of tumors toward the prostate boundary and the proportions of slides with prostate cancer. Gleason pattern 4 was particularly associated with discordance. Notably, grade discordance with vPatho was not specific to any of the six pathologists involved in routine clinical grading. In conclusion, our study highlights the potential utility of AI in developing a digital twin of a pathologist. This approach can help uncover limitations in AI adoption and the current grading system for prostate cancer pathology.
Conceptual Framework and Documentation Standards of Cystoscopic Media Content for Artificial Intelligence
Jan 18, 2023



Abstract:Background: The clinical documentation of cystoscopy includes visual and textual materials. However, the secondary use of visual cystoscopic data for educational and research purposes remains limited due to inefficient data management in routine clinical practice. Methods: A conceptual framework was designed to document cystoscopy in a standardized manner with three major sections: data management, annotation management, and utilization management. A Swiss-cheese model was proposed for quality control and root cause analyses. We defined the infrastructure required to implement the framework with respect to FAIR (findable, accessible, interoperable, re-usable) principles. We applied two scenarios exemplifying data sharing for research and educational projects to ensure the compliance with FAIR principles. Results: The framework was successfully implemented while following FAIR principles. The cystoscopy atlas produced from the framework could be presented in an educational web portal; a total of 68 full-length qualitative videos and corresponding annotation data were sharable for artificial intelligence projects covering frame classification and segmentation problems at case, lesion and frame levels. Conclusion: Our study shows that the proposed framework facilitates the storage of the visual documentation in a standardized manner and enables FAIR data for education and artificial intelligence research.
CheXstray: Real-time Multi-Modal Data Concordance for Drift Detection in Medical Imaging AI
Feb 06, 2022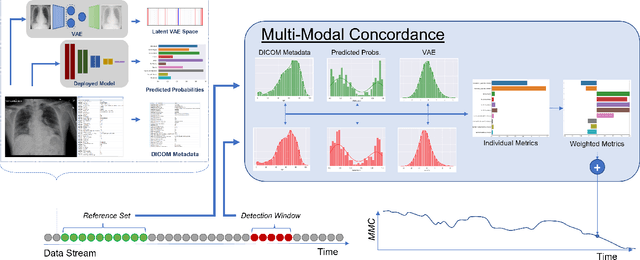
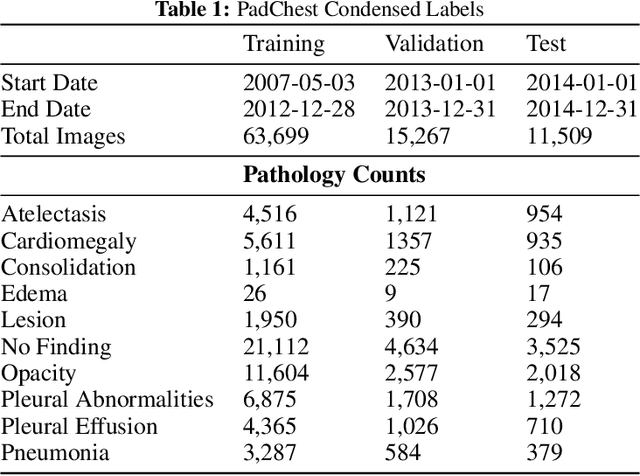
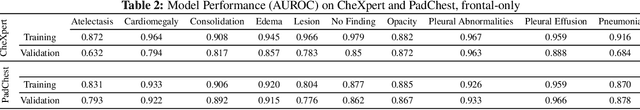
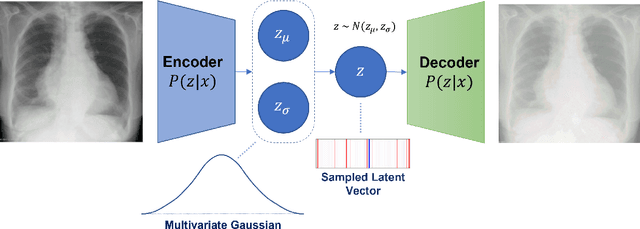
Abstract:Rapidly expanding Clinical AI applications worldwide have the potential to impact to all areas of medical practice. Medical imaging applications constitute a vast majority of approved clinical AI applications. Though healthcare systems are eager to adopt AI solutions a fundamental question remains: \textit{what happens after the AI model goes into production?} We use the CheXpert and PadChest public datasets to build and test a medical imaging AI drift monitoring workflow that tracks data and model drift without contemporaneous ground truth. We simulate drift in multiple experiments to compare model performance with our novel multi-modal drift metric, which uses DICOM metadata, image appearance representation from a variational autoencoder (VAE), and model output probabilities as input. Through experimentation, we demonstrate a strong proxy for ground truth performance using unsupervised distributional shifts in relevant metadata, predicted probabilities, and VAE latent representation. Our key contributions include (1) proof-of-concept for medical imaging drift detection including use of VAE and domain specific statistical methods (2) a multi-modal methodology for measuring and unifying drift metrics (3) new insights into the challenges and solutions for observing deployed medical imaging AI (4) creation of open-source tools enabling others to easily run their own workflows or scenarios. This work has important implications for addressing the translation gap related to continuous medical imaging AI model monitoring in dynamic healthcare environments.
CheXbreak: Misclassification Identification for Deep Learning Models Interpreting Chest X-rays
Mar 24, 2021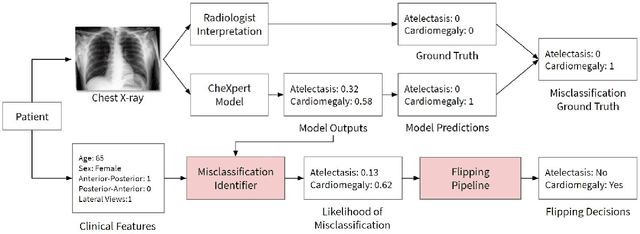
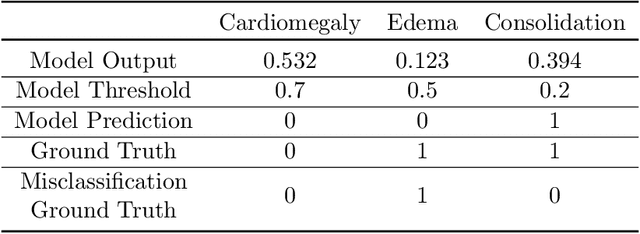
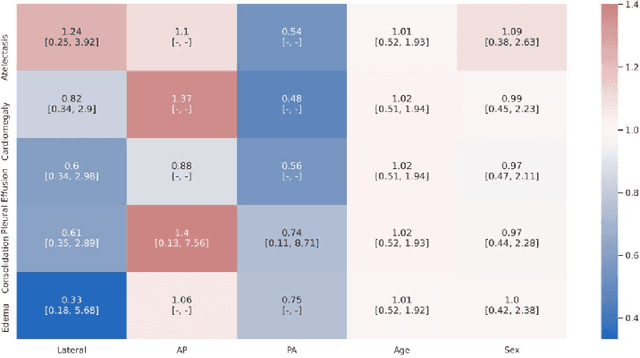
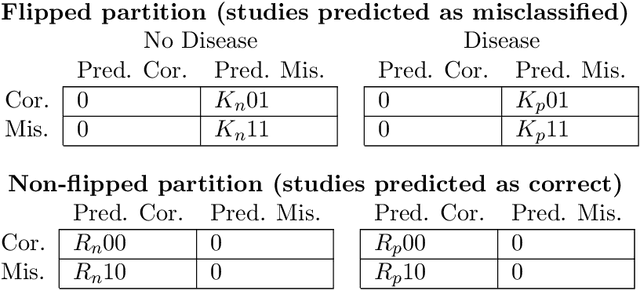
Abstract:A major obstacle to the integration of deep learning models for chest x-ray interpretation into clinical settings is the lack of understanding of their failure modes. In this work, we first investigate whether there are patient subgroups that chest x-ray models are likely to misclassify. We find that patient age and the radiographic finding of lung lesion, pneumothorax or support devices are statistically relevant features for predicting misclassification for some chest x-ray models. Second, we develop misclassification predictors on chest x-ray models using their outputs and clinical features. We find that our best performing misclassification identifier achieves an AUROC close to 0.9 for most diseases. Third, employing our misclassification identifiers, we develop a corrective algorithm to selectively flip model predictions that have high likelihood of misclassification at inference time. We observe F1 improvement on the prediction of Consolidation (0.008 [95\% CI 0.005, 0.010]) and Edema (0.003, [95\% CI 0.001, 0.006]). By carrying out our investigation on ten distinct and high-performing chest x-ray models, we are able to derive insights across model architectures and offer a generalizable framework applicable to other medical imaging tasks.
Learning domain-agnostic visual representation for computational pathology using medically-irrelevant style transfer augmentation
Feb 02, 2021



Abstract:Suboptimal generalization of machine learning models on unseen data is a key challenge which hampers the clinical applicability of such models to medical imaging. Although various methods such as domain adaptation and domain generalization have evolved to combat this challenge, learning robust and generalizable representations is core to medical image understanding, and continues to be a problem. Here, we propose STRAP (Style TRansfer Augmentation for histoPathology), a form of data augmentation based on random style transfer from artistic paintings, for learning domain-agnostic visual representations in computational pathology. Style transfer replaces the low-level texture content of images with the uninformative style of randomly selected artistic paintings, while preserving high-level semantic content. This improves robustness to domain shift and can be used as a simple yet powerful tool for learning domain-agnostic representations. We demonstrate that STRAP leads to state-of-the-art performance, particularly in the presence of domain shifts, on a particular classification task of predicting microsatellite status in colorectal cancer using digitized histopathology images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge