James D. Brooks
Critical Evaluation of Artificial Intelligence as Digital Twin of Pathologist for Prostate Cancer Pathology
Aug 23, 2023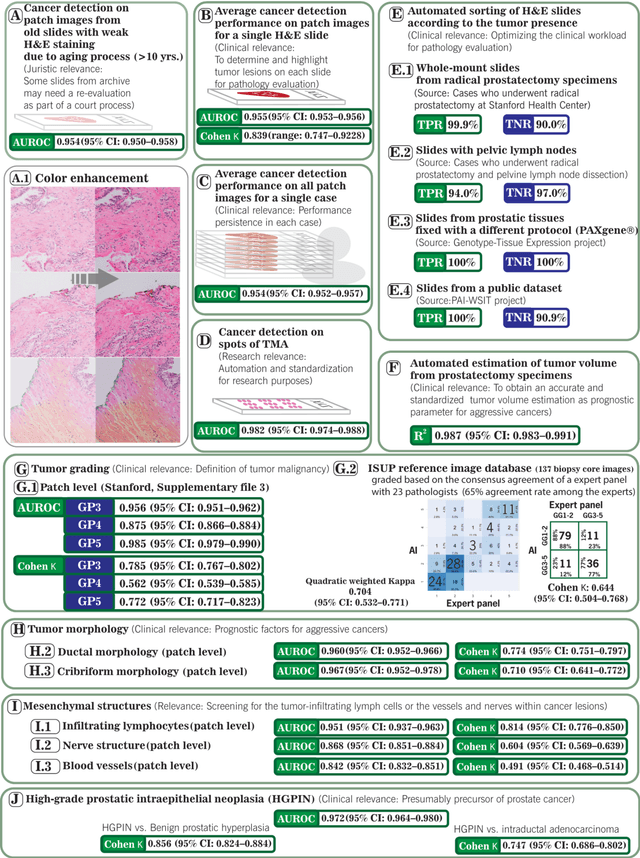
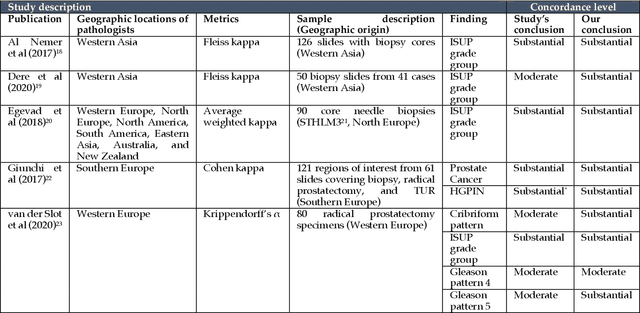
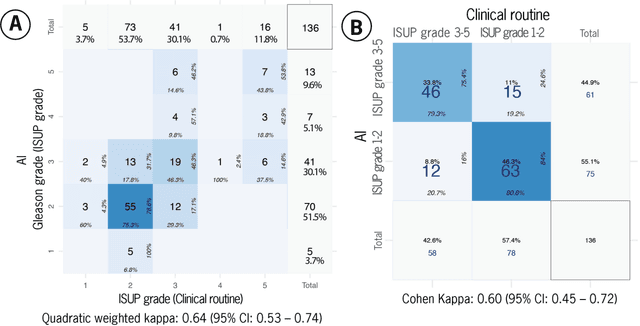
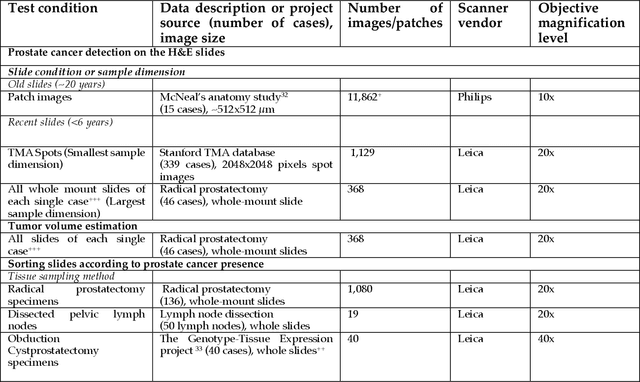
Abstract:Prostate cancer pathology plays a crucial role in clinical management but is time-consuming. Artificial intelligence (AI) shows promise in detecting prostate cancer and grading patterns. We tested an AI-based digital twin of a pathologist, vPatho, on 2,603 histology images of prostate tissue stained with hematoxylin and eosin. We analyzed various factors influencing tumor-grade disagreement between vPatho and six human pathologists. Our results demonstrated that vPatho achieved comparable performance in prostate cancer detection and tumor volume estimation, as reported in the literature. Concordance levels between vPatho and human pathologists were examined. Notably, moderate to substantial agreement was observed in identifying complementary histological features such as ductal, cribriform, nerve, blood vessels, and lymph cell infiltrations. However, concordance in tumor grading showed a decline when applied to prostatectomy specimens (kappa = 0.44) compared to biopsy cores (kappa = 0.70). Adjusting the decision threshold for the secondary Gleason pattern from 5% to 10% improved the concordance level between pathologists and vPatho for tumor grading on prostatectomy specimens (kappa from 0.44 to 0.64). Potential causes of grade discordance included the vertical extent of tumors toward the prostate boundary and the proportions of slides with prostate cancer. Gleason pattern 4 was particularly associated with discordance. Notably, grade discordance with vPatho was not specific to any of the six pathologists involved in routine clinical grading. In conclusion, our study highlights the potential utility of AI in developing a digital twin of a pathologist. This approach can help uncover limitations in AI adoption and the current grading system for prostate cancer pathology.
Correlated Feature Aggregation by Region Helps Distinguish Aggressive from Indolent Clear Cell Renal Cell Carcinoma Subtypes on CT
Sep 29, 2022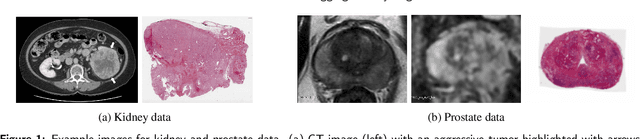
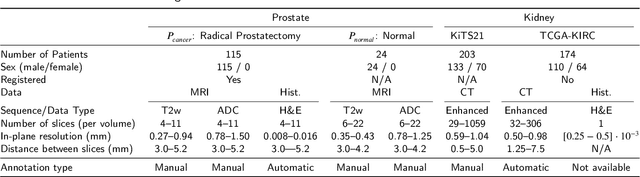
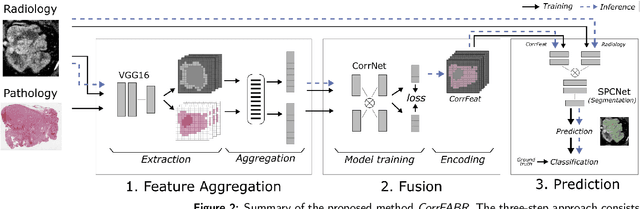

Abstract:Renal cell carcinoma (RCC) is a common cancer that varies in clinical behavior. Indolent RCC is often low-grade without necrosis and can be monitored without treatment. Aggressive RCC is often high-grade and can cause metastasis and death if not promptly detected and treated. While most kidney cancers are detected on CT scans, grading is based on histology from invasive biopsy or surgery. Determining aggressiveness on CT images is clinically important as it facilitates risk stratification and treatment planning. This study aims to use machine learning methods to identify radiology features that correlate with features on pathology to facilitate assessment of cancer aggressiveness on CT images instead of histology. This paper presents a novel automated method, Correlated Feature Aggregation By Region (CorrFABR), for classifying aggressiveness of clear cell RCC by leveraging correlations between radiology and corresponding unaligned pathology images. CorrFABR consists of three main steps: (1) Feature Aggregation where region-level features are extracted from radiology and pathology images, (2) Fusion where radiology features correlated with pathology features are learned on a region level, and (3) Prediction where the learned correlated features are used to distinguish aggressive from indolent clear cell RCC using CT alone as input. Thus, during training, CorrFABR learns from both radiology and pathology images, but during inference, CorrFABR will distinguish aggressive from indolent clear cell RCC using CT alone, in the absence of pathology images. CorrFABR improved classification performance over radiology features alone, with an increase in binary classification F1-score from 0.68 (0.04) to 0.73 (0.03). This demonstrates the potential of incorporating pathology disease characteristics for improved classification of aggressiveness of clear cell RCC on CT images.
Bridging the gap between prostate radiology and pathology through machine learning
Dec 03, 2021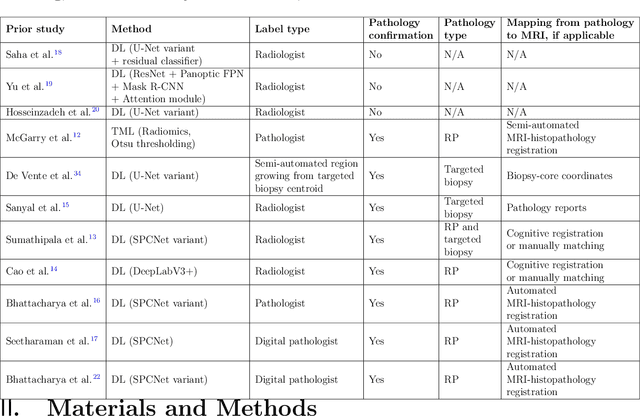
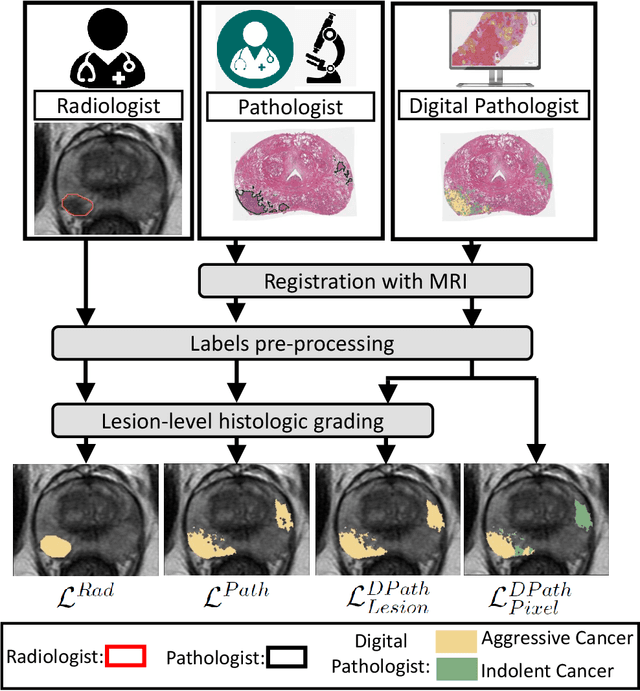
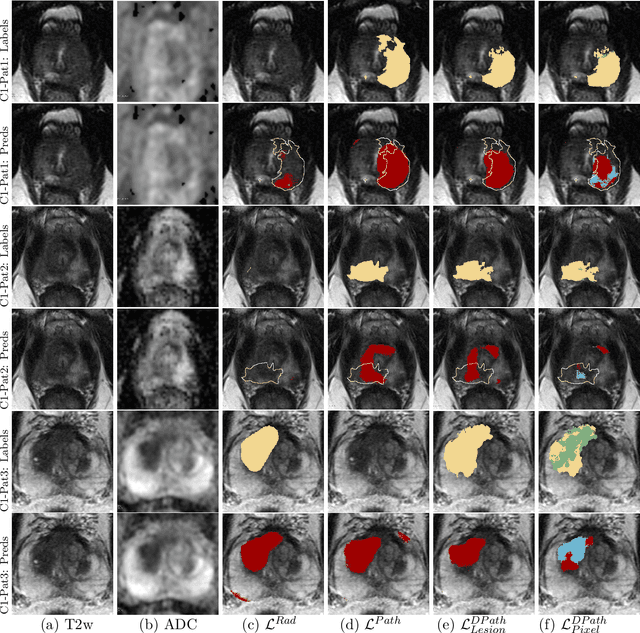
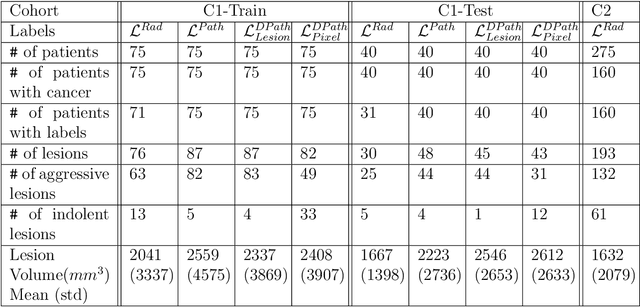
Abstract:Prostate cancer is the second deadliest cancer for American men. While Magnetic Resonance Imaging (MRI) is increasingly used to guide targeted biopsies for prostate cancer diagnosis, its utility remains limited due to high rates of false positives and false negatives as well as low inter-reader agreements. Machine learning methods to detect and localize cancer on prostate MRI can help standardize radiologist interpretations. However, existing machine learning methods vary not only in model architecture, but also in the ground truth labeling strategies used for model training. In this study, we compare different labeling strategies, namely, pathology-confirmed radiologist labels, pathologist labels on whole-mount histopathology images, and lesion-level and pixel-level digital pathologist labels (previously validated deep learning algorithm on histopathology images to predict pixel-level Gleason patterns) on whole-mount histopathology images. We analyse the effects these labels have on the performance of the trained machine learning models. Our experiments show that (1) radiologist labels and models trained with them can miss cancers, or underestimate cancer extent, (2) digital pathologist labels and models trained with them have high concordance with pathologist labels, and (3) models trained with digital pathologist labels achieve the best performance in prostate cancer detection in two different cohorts with different disease distributions, irrespective of the model architecture used. Digital pathologist labels can reduce challenges associated with human annotations, including labor, time, inter- and intra-reader variability, and can help bridge the gap between prostate radiology and pathology by enabling the training of reliable machine learning models to detect and localize prostate cancer on MRI.
Weakly Supervised Registration of Prostate MRI and Histopathology Images
Jun 23, 2021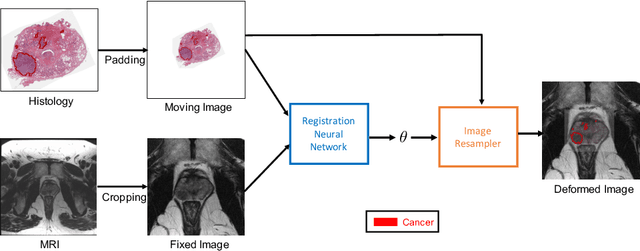
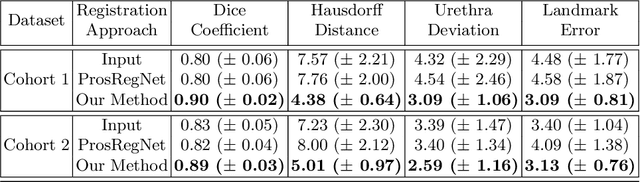
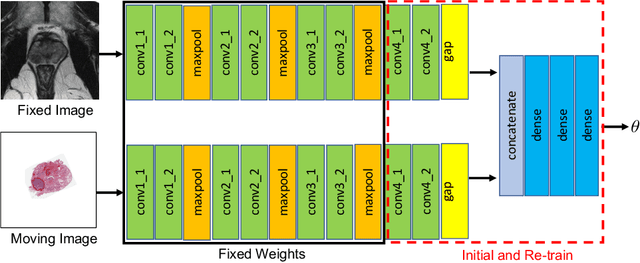
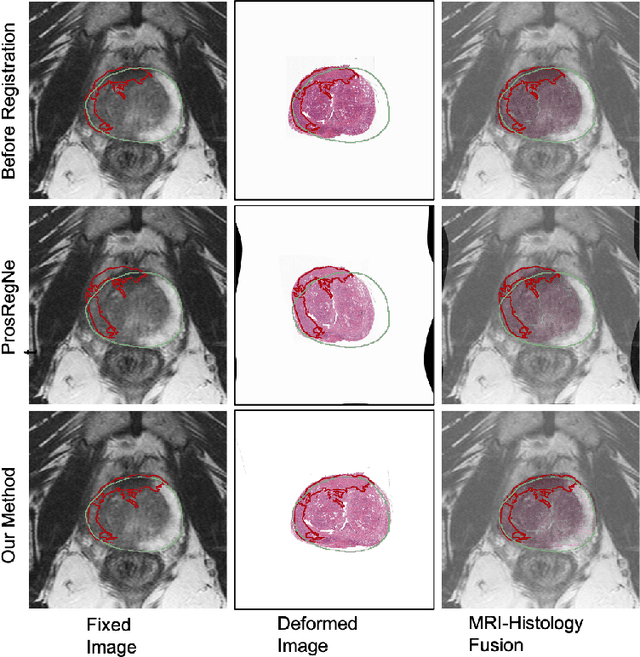
Abstract:The interpretation of prostate MRI suffers from low agreement across radiologists due to the subtle differences between cancer and normal tissue. Image registration addresses this issue by accurately mapping the ground-truth cancer labels from surgical histopathology images onto MRI. Cancer labels achieved by image registration can be used to improve radiologists' interpretation of MRI by training deep learning models for early detection of prostate cancer. A major limitation of current automated registration approaches is that they require manual prostate segmentations, which is a time-consuming task, prone to errors. This paper presents a weakly supervised approach for affine and deformable registration of MRI and histopathology images without requiring prostate segmentations. We used manual prostate segmentations and mono-modal synthetic image pairs to train our registration networks to align prostate boundaries and local prostate features. Although prostate segmentations were used during the training of the network, such segmentations were not needed when registering unseen images at inference time. We trained and validated our registration network with 135 and 10 patients from an internal cohort, respectively. We tested the performance of our method using 16 patients from the internal cohort and 22 patients from an external cohort. The results show that our weakly supervised method has achieved significantly higher registration accuracy than a state-of-the-art method run without prostate segmentations. Our deep learning framework will ease the registration of MRI and histopathology images by obviating the need for prostate segmentations.
CorrSigNet: Learning CORRelated Prostate Cancer SIGnatures from Radiology and Pathology Images for Improved Computer Aided Diagnosis
Jul 31, 2020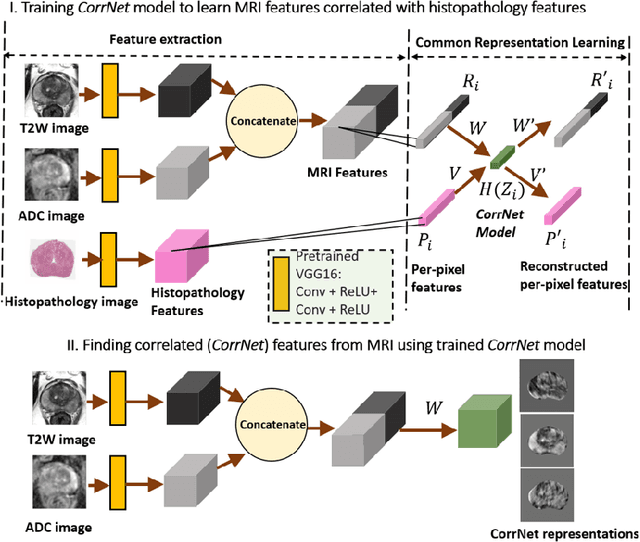
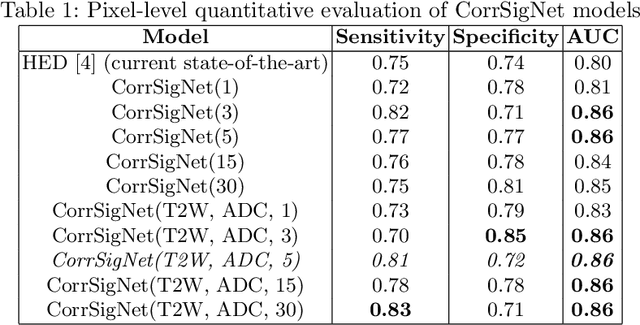
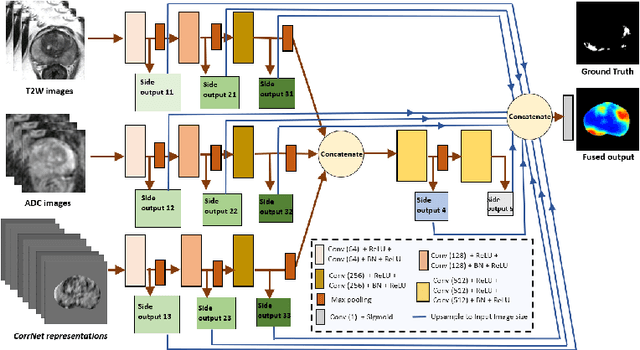
Abstract:Magnetic Resonance Imaging (MRI) is widely used for screening and staging prostate cancer. However, many prostate cancers have subtle features which are not easily identifiable on MRI, resulting in missed diagnoses and alarming variability in radiologist interpretation. Machine learning models have been developed in an effort to improve cancer identification, but current models localize cancer using MRI-derived features, while failing to consider the disease pathology characteristics observed on resected tissue. In this paper, we propose CorrSigNet, an automated two-step model that localizes prostate cancer on MRI by capturing the pathology features of cancer. First, the model learns MRI signatures of cancer that are correlated with corresponding histopathology features using Common Representation Learning. Second, the model uses the learned correlated MRI features to train a Convolutional Neural Network to localize prostate cancer. The histopathology images are used only in the first step to learn the correlated features. Once learned, these correlated features can be extracted from MRI of new patients (without histopathology or surgery) to localize cancer. We trained and validated our framework on a unique dataset of 75 patients with 806 slices who underwent MRI followed by prostatectomy surgery. We tested our method on an independent test set of 20 prostatectomy patients (139 slices, 24 cancerous lesions, 1.12M pixels) and achieved a per-pixel sensitivity of 0.81, specificity of 0.71, AUC of 0.86 and a per-lesion AUC of $0.96 \pm 0.07$, outperforming the current state-of-the-art accuracy in predicting prostate cancer using MRI.
Plexus Convolutional Neural Network (PlexusNet): A novel neural network architecture for histologic image analysis
Aug 24, 2019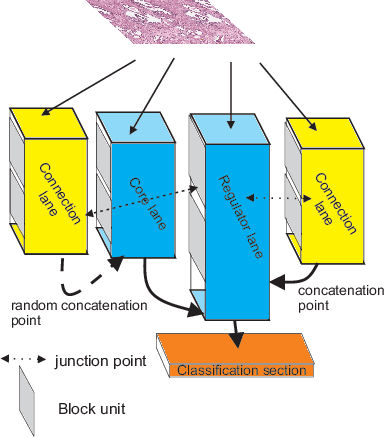
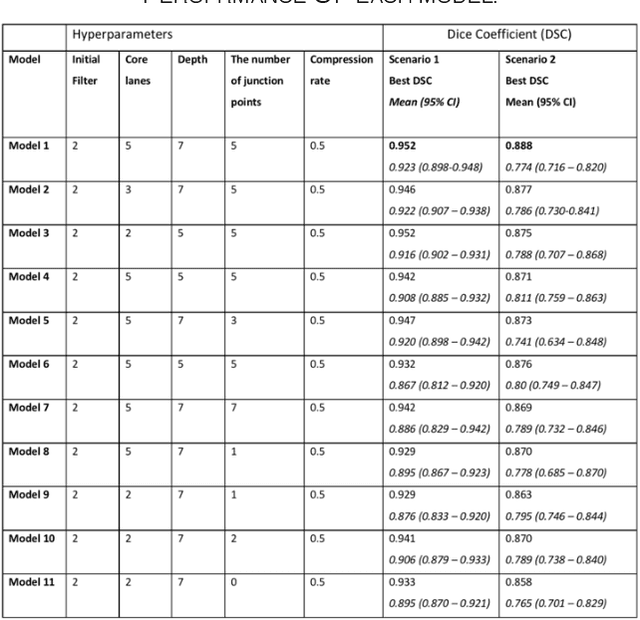
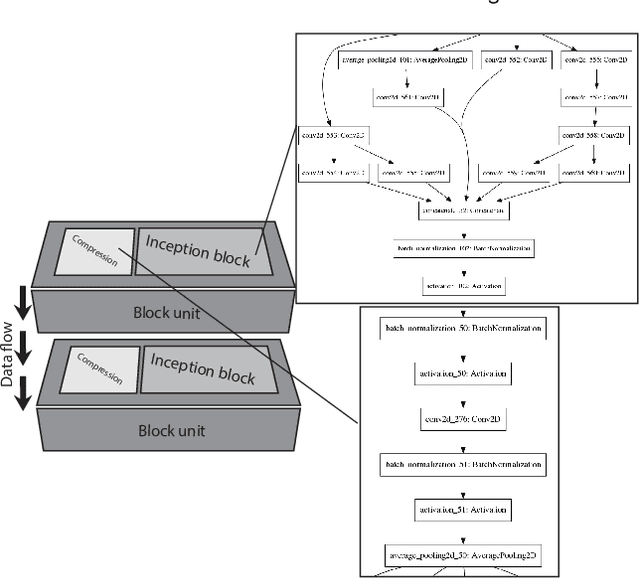
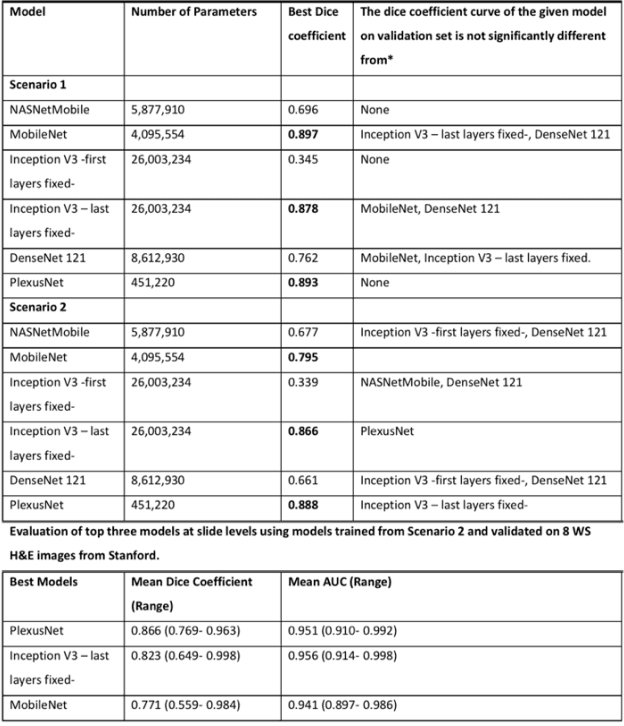
Abstract:Different convolutional neural network (CNN) models have been tested for their application in histologic imaging analyses. However, these models are prone to overfitting due to their large parameter capacity, requiring more data and expensive computational resources for model training. Given these limitations, we developed and tested PlexusNet for histologic evaluation using a single GPU by a batch dimension of 16x512x512x3. We utilized 62 Hematoxylin and eosin stain (H&E) annotated histological images of radical prostatectomy cases from TCGA-PRAD and Stanford University, and 24 H&E whole-slide images with hepatocellular carcinoma from TCGA-LIHC diagnostic histology images. Base models were DenseNet, Inception V3, and MobileNet and compared with PlexusNet. The dice coefficient (DSC) was evaluated for each model. PlexusNet delivered comparable classification performance (DSC at patch level: 0.89) for H&E whole-slice images in distinguishing prostate cancer from normal tissues. The parameter capacity of PlexusNet is 9 times smaller than MobileNet or 58 times smaller than Inception V3, respectively. Similar findings were observed in distinguishing hepatocellular carcinoma from non-cancerous liver histologies (DSC at patch level: 0.85). As conclusion, PlexusNet represents a novel model architecture for histological image analysis that achieves classification performance comparable to the base models while providing orders-of-magnitude memory savings.
A Historical Analysis of the Field of OR/MS using Topic Models
Oct 17, 2015
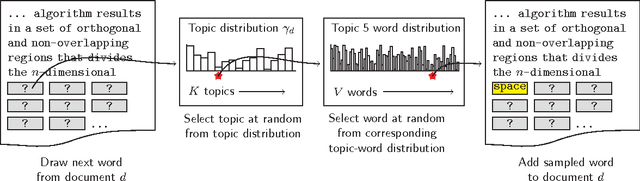
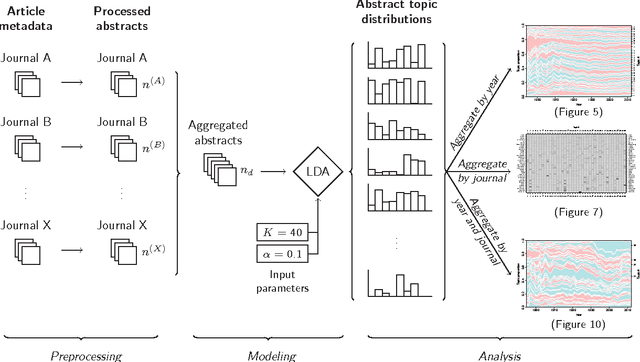
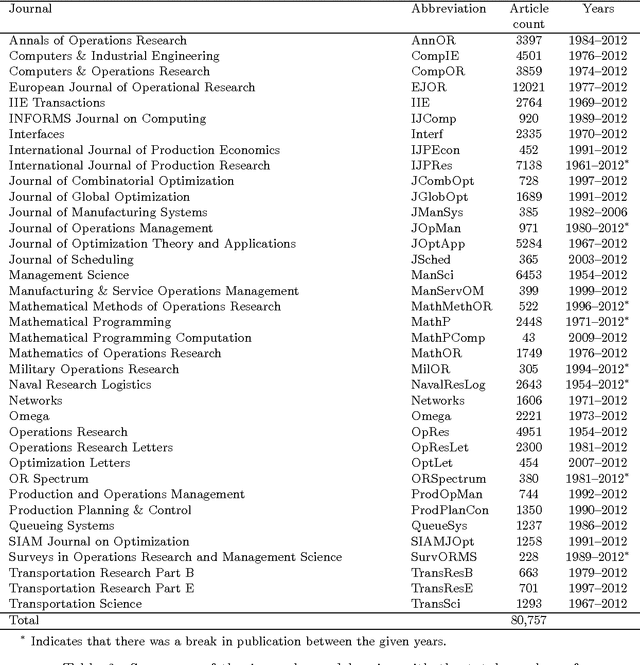
Abstract:This study investigates the content of the published scientific literature in the fields of operations research and management science (OR/MS) since the early 1950s. Our study is based on 80,757 published journal abstracts from 37 of the leading OR/MS journals. We have developed a topic model, using Latent Dirichlet Allocation (LDA), and extend this analysis to reveal the temporal dynamics of the field, journals, and topics. Our analysis shows the generality or specificity of each of the journals, and we identify groups of journals with similar content, which are both consistent and inconsistent with intuition. We also show how journals have become more or less unique in their scope. A more detailed analysis of each journals' topics over time shows significant temporal dynamics, especially for journals with niche content. This study presents an observational, yet objective, view of the published literature from OR/MS that would be of interest to authors, editors, journals, and publishers. Furthermore, this work can be used by new entrants to the fields of OR/MS to understand the content landscape, as a starting point for discussions and inquiry of the field at large, or as a model for other fields to perform similar analyses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge