Rewa Sood
CorrSigNet: Learning CORRelated Prostate Cancer SIGnatures from Radiology and Pathology Images for Improved Computer Aided Diagnosis
Jul 31, 2020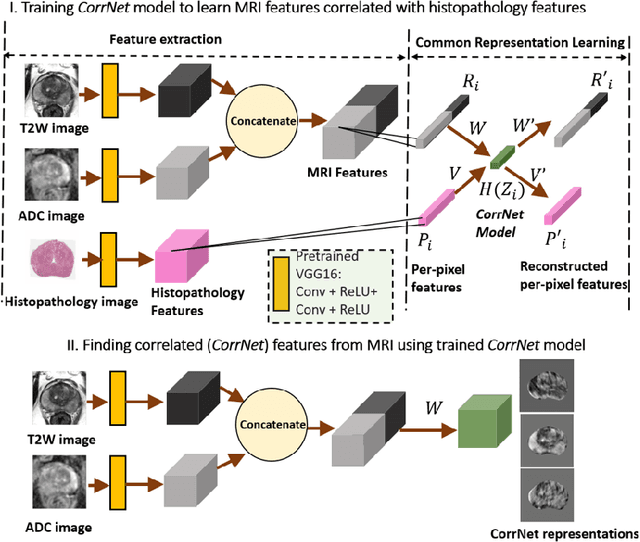
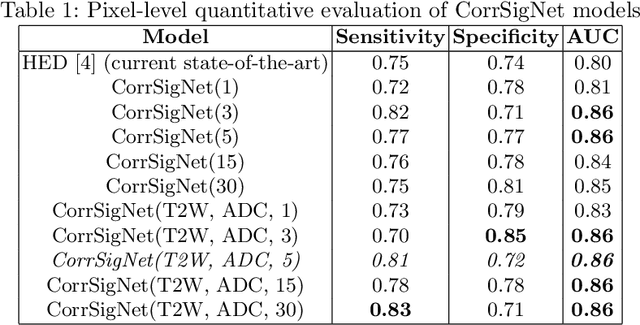
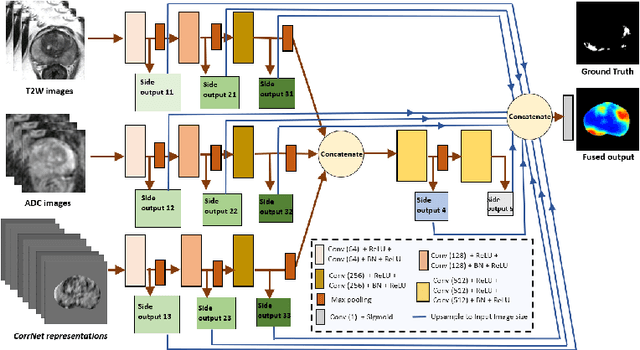
Abstract:Magnetic Resonance Imaging (MRI) is widely used for screening and staging prostate cancer. However, many prostate cancers have subtle features which are not easily identifiable on MRI, resulting in missed diagnoses and alarming variability in radiologist interpretation. Machine learning models have been developed in an effort to improve cancer identification, but current models localize cancer using MRI-derived features, while failing to consider the disease pathology characteristics observed on resected tissue. In this paper, we propose CorrSigNet, an automated two-step model that localizes prostate cancer on MRI by capturing the pathology features of cancer. First, the model learns MRI signatures of cancer that are correlated with corresponding histopathology features using Common Representation Learning. Second, the model uses the learned correlated MRI features to train a Convolutional Neural Network to localize prostate cancer. The histopathology images are used only in the first step to learn the correlated features. Once learned, these correlated features can be extracted from MRI of new patients (without histopathology or surgery) to localize cancer. We trained and validated our framework on a unique dataset of 75 patients with 806 slices who underwent MRI followed by prostatectomy surgery. We tested our method on an independent test set of 20 prostatectomy patients (139 slices, 24 cancerous lesions, 1.12M pixels) and achieved a per-pixel sensitivity of 0.81, specificity of 0.71, AUC of 0.86 and a per-lesion AUC of $0.96 \pm 0.07$, outperforming the current state-of-the-art accuracy in predicting prostate cancer using MRI.
An Application of Generative Adversarial Networks for Super Resolution Medical Imaging
Dec 19, 2019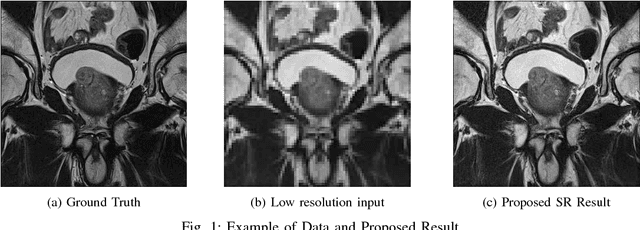
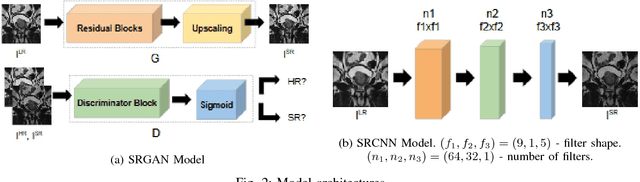

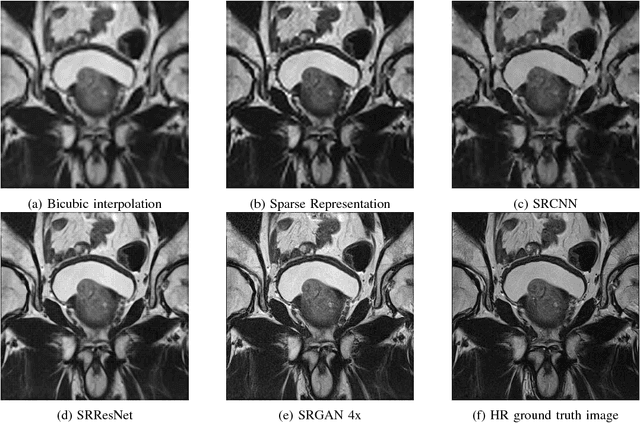
Abstract:Acquiring High Resolution (HR) Magnetic Resonance (MR) images requires the patient to remain still for long periods of time, which causes patient discomfort and increases the probability of motion induced image artifacts. A possible solution is to acquire low resolution (LR) images and to process them with the Super Resolution Generative Adversarial Network (SRGAN) to create an HR version. Acquiring LR images requires a lower scan time than acquiring HR images, which allows for higher patient comfort and scanner throughput. This work applies SRGAN to MR images of the prostate to improve the in-plane resolution by factors of 4 and 8. The term 'super resolution' in the context of this paper defines the post processing enhancement of medical images as opposed to 'high resolution' which defines native image resolution acquired during the MR acquisition phase. We also compare the SRGAN to three other models: SRCNN, SRResNet, and Sparse Representation. While the SRGAN results do not have the best Peak Signal to Noise Ratio (PSNR) or Structural Similarity (SSIM) metrics, they are the visually most similar to the original HR images, as portrayed by the Mean Opinion Score (MOS) results.
* International Conference on Machine Learning Applications, 6 pages, 5 figures, 2 tables
Anisotropic Super Resolution in Prostate MRI using Super Resolution Generative Adversarial Networks
Dec 19, 2019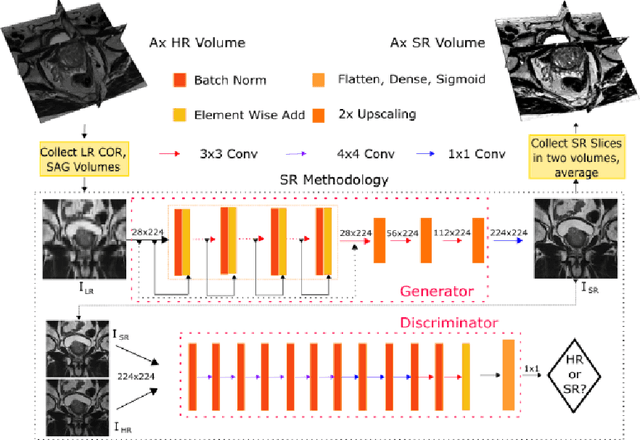
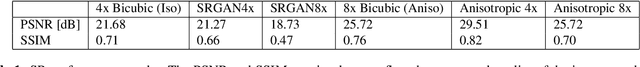
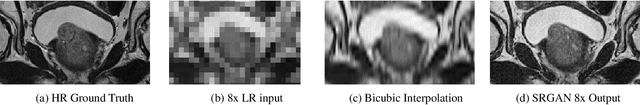
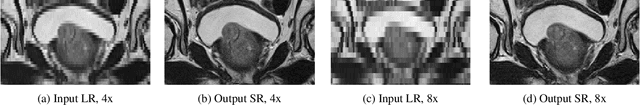
Abstract:Acquiring High Resolution (HR) Magnetic Resonance (MR) images requires the patient to remain still for long periods of time, which causes patient discomfort and increases the probability of motion induced image artifacts. A possible solution is to acquire low resolution (LR) images and to process them with the Super Resolution Generative Adversarial Network (SRGAN) to create a super-resolved version. This work applies SRGAN to MR images of the prostate and performs three experiments. The first experiment explores improving the in-plane MR image resolution by factors of 4 and 8, and shows that, while the PSNR and SSIM (Structural SIMilarity) metrics are lower than the isotropic bicubic interpolation baseline, the SRGAN is able to create images that have high edge fidelity. The second experiment explores anisotropic super-resolution via synthetic images, in that the input images to the network are anisotropically downsampled versions of HR images. This experiment demonstrates the ability of the modified SRGAN to perform anisotropic super-resolution, with quantitative image metrics that are comparable to those of the anisotropic bicubic interpolation baseline. Finally, the third experiment applies a modified version of the SRGAN to super-resolve anisotropic images obtained from the through-plane slices of the volumetric MR data. The output super-resolved images contain a significant amount of high frequency information that make them visually close to their HR counterparts. Overall, the promising results from each experiment show that super-resolution for MR images is a successful technique and that producing isotropic MR image volumes from anisotropic slices is an achievable goal.
* International Symposium on Biomedical Imaging, 4 pages, 4 figures, 1 table
BERTQA -- Attention on Steroids
Dec 14, 2019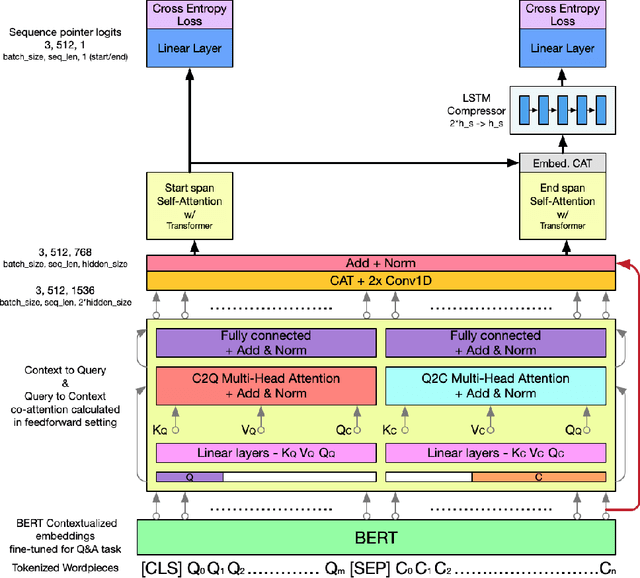
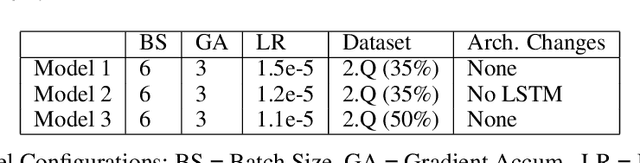
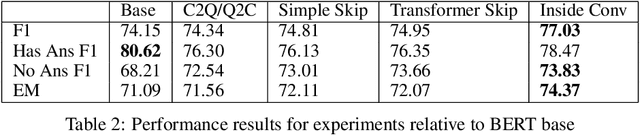
Abstract:In this work, we extend the Bidirectional Encoder Representations from Transformers (BERT) with an emphasis on directed coattention to obtain an improved F1 performance on the SQUAD2.0 dataset. The Transformer architecture on which BERT is based places hierarchical global attention on the concatenation of the context and query. Our additions to the BERT architecture augment this attention with a more focused context to query (C2Q) and query to context (Q2C) attention via a set of modified Transformer encoder units. In addition, we explore adding convolution-based feature extraction within the coattention architecture to add localized information to self-attention. We found that coattention significantly improves the no answer F1 by 4 points in the base and 1 point in the large architecture. After adding skip connections the no answer F1 improved further without causing an additional loss in has answer F1. The addition of localized feature extraction added to attention produced an overall dev F1 of 77.03 in the base architecture. We applied our findings to the large BERT model which contains twice as many layers and further used our own augmented version of the SQUAD 2.0 dataset created by back translation, which we have named SQUAD 2.Q. Finally, we performed hyperparameter tuning and ensembled our best models for a final F1/EM of 82.317/79.442 (Attention on Steroids, PCE Test Leaderboard).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge