Ricardo Montoya-del-Angel
Tumor Detection, Segmentation and Classification Challenge on Automated 3D Breast Ultrasound: The TDSC-ABUS Challenge
Jan 26, 2025



Abstract:Breast cancer is one of the most common causes of death among women worldwide. Early detection helps in reducing the number of deaths. Automated 3D Breast Ultrasound (ABUS) is a newer approach for breast screening, which has many advantages over handheld mammography such as safety, speed, and higher detection rate of breast cancer. Tumor detection, segmentation, and classification are key components in the analysis of medical images, especially challenging in the context of 3D ABUS due to the significant variability in tumor size and shape, unclear tumor boundaries, and a low signal-to-noise ratio. The lack of publicly accessible, well-labeled ABUS datasets further hinders the advancement of systems for breast tumor analysis. Addressing this gap, we have organized the inaugural Tumor Detection, Segmentation, and Classification Challenge on Automated 3D Breast Ultrasound 2023 (TDSC-ABUS2023). This initiative aims to spearhead research in this field and create a definitive benchmark for tasks associated with 3D ABUS image analysis. In this paper, we summarize the top-performing algorithms from the challenge and provide critical analysis for ABUS image examination. We offer the TDSC-ABUS challenge as an open-access platform at https://tdsc-abus2023.grand-challenge.org/ to benchmark and inspire future developments in algorithmic research.
CoMoTo: Unpaired Cross-Modal Lesion Distillation Improves Breast Lesion Detection in Tomosynthesis
Jul 24, 2024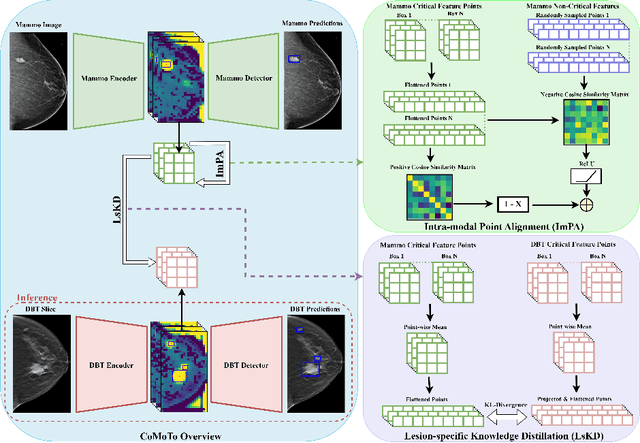
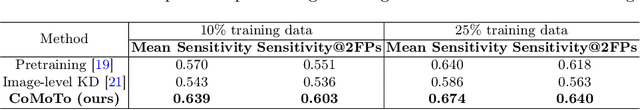
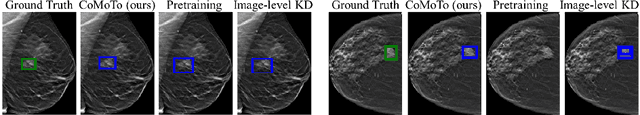
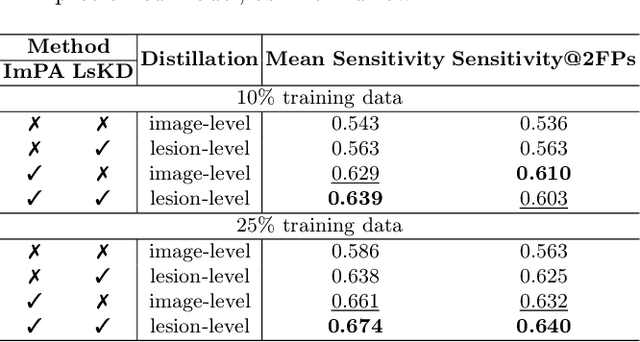
Abstract:Digital Breast Tomosynthesis (DBT) is an advanced breast imaging modality that offers superior lesion detection accuracy compared to conventional mammography, albeit at the trade-off of longer reading time. Accelerating lesion detection from DBT using deep learning is hindered by limited data availability and huge annotation costs. A possible solution to this issue could be to leverage the information provided by a more widely available modality, such as mammography, to enhance DBT lesion detection. In this paper, we present a novel framework, CoMoTo, for improving lesion detection in DBT. Our framework leverages unpaired mammography data to enhance the training of a DBT model, improving practicality by eliminating the need for mammography during inference. Specifically, we propose two novel components, Lesion-specific Knowledge Distillation (LsKD) and Intra-modal Point Alignment (ImPA). LsKD selectively distills lesion features from a mammography teacher model to a DBT student model, disregarding background features. ImPA further enriches LsKD by ensuring the alignment of lesion features within the teacher before distilling knowledge to the student. Our comprehensive evaluation shows that CoMoTo is superior to traditional pretraining and image-level KD, improving performance by 7% Mean Sensitivity under low-data setting. Our code is available at https://github.com/Muhammad-Al-Barbary/CoMoTo .
MAM-E: Mammographic synthetic image generation with diffusion models
Nov 16, 2023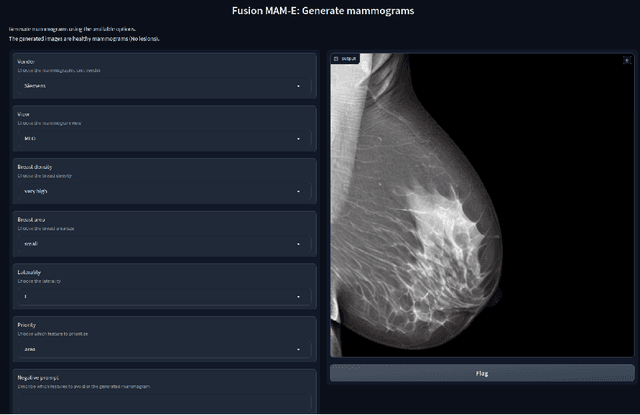


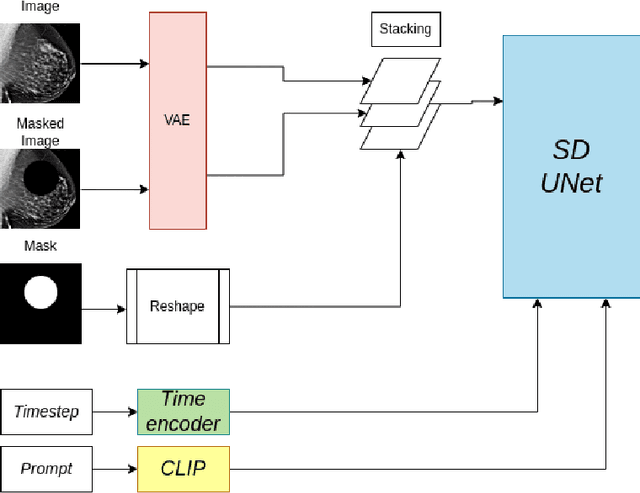
Abstract:Generative models are used as an alternative data augmentation technique to alleviate the data scarcity problem faced in the medical imaging field. Diffusion models have gathered special attention due to their innovative generation approach, the high quality of the generated images and their relatively less complex training process compared with Generative Adversarial Networks. Still, the implementation of such models in the medical domain remains at early stages. In this work, we propose exploring the use of diffusion models for the generation of high quality full-field digital mammograms using state-of-the-art conditional diffusion pipelines. Additionally, we propose using stable diffusion models for the inpainting of synthetic lesions on healthy mammograms. We introduce MAM-E, a pipeline of generative models for high quality mammography synthesis controlled by a text prompt and capable of generating synthetic lesions on specific regions of the breast. Finally, we provide quantitative and qualitative assessment of the generated images and easy-to-use graphical user interfaces for mammography synthesis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge