Eloy García
MRI Breast tissue segmentation using nnU-Net for biomechanical modeling
Nov 27, 2024Abstract:Integrating 2D mammography with 3D magnetic resonance imaging (MRI) is crucial for improving breast cancer diagnosis and treatment planning. However, this integration is challenging due to differences in imaging modalities and the need for precise tissue segmentation and alignment. This paper addresses these challenges by enhancing biomechanical breast models in two main aspects: improving tissue identification using nnU-Net segmentation models and evaluating finite element (FE) biomechanical solvers, specifically comparing NiftySim and FEBio. We performed a detailed six-class segmentation of breast MRI data using the nnU-Net architecture, achieving Dice Coefficients of 0.94 for fat, 0.88 for glandular tissue, and 0.87 for pectoral muscle. The overall foreground segmentation reached a mean Dice Coefficient of 0.83 through an ensemble of 2D and 3D U-Net configurations, providing a solid foundation for 3D reconstruction and biomechanical modeling. The segmented data was then used to generate detailed 3D meshes and develop biomechanical models using NiftySim and FEBio, which simulate breast tissue's physical behaviors under compression. Our results include a comparison between NiftySim and FEBio, providing insights into the accuracy and reliability of these simulations in studying breast tissue responses under compression. The findings of this study have the potential to improve the integration of 2D and 3D imaging modalities, thereby enhancing diagnostic accuracy and treatment planning for breast cancer.
Graph Neural Networks for modelling breast biomechanical compression
Nov 10, 2024



Abstract:Breast compression simulation is essential for accurate image registration from 3D modalities to X-ray procedures like mammography. It accounts for tissue shape and position changes due to compression, ensuring precise alignment and improved analysis. Although Finite Element Analysis (FEA) is reliable for approximating soft tissue deformation, it struggles with balancing accuracy and computational efficiency. Recent studies have used data-driven models trained on FEA results to speed up tissue deformation predictions. We propose to explore Physics-based Graph Neural Networks (PhysGNN) for breast compression simulation. PhysGNN has been used for data-driven modelling in other domains, and this work presents the first investigation of their potential in predicting breast deformation during mammographic compression. Unlike conventional data-driven models, PhysGNN, which incorporates mesh structural information and enables inductive learning on unstructured grids, is well-suited for capturing complex breast tissue geometries. Trained on deformations from incremental FEA simulations, PhysGNN's performance is evaluated by comparing predicted nodal displacements with those from finite element (FE) simulations. This deep learning (DL) framework shows promise for accurate, rapid breast deformation approximations, offering enhanced computational efficiency for real-world scenarios.
medigan: A Python Library of Pretrained Generative Models for Enriched Data Access in Medical Imaging
Sep 28, 2022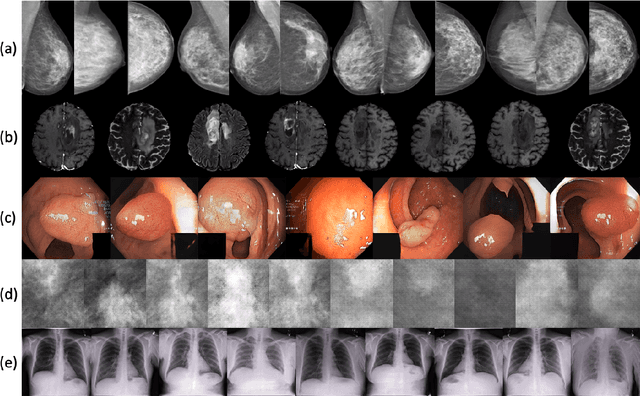
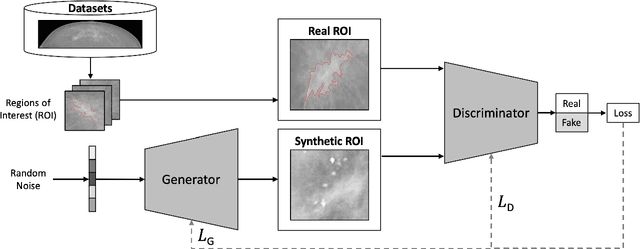
Abstract:Synthetic data generated by generative models can enhance the performance and capabilities of data-hungry deep learning models in medical imaging. However, there is (1) limited availability of (synthetic) datasets and (2) generative models are complex to train, which hinders their adoption in research and clinical applications. To reduce this entry barrier, we propose medigan, a one-stop shop for pretrained generative models implemented as an open-source framework-agnostic Python library. medigan allows researchers and developers to create, increase, and domain-adapt their training data in just a few lines of code. Guided by design decisions based on gathered end-user requirements, we implement medigan based on modular components for generative model (i) execution, (ii) visualisation, (iii) search & ranking, and (iv) contribution. The library's scalability and design is demonstrated by its growing number of integrated and readily-usable pretrained generative models consisting of 21 models utilising 9 different Generative Adversarial Network architectures trained on 11 datasets from 4 domains, namely, mammography, endoscopy, x-ray, and MRI. Furthermore, 3 applications of medigan are analysed in this work, which include (a) enabling community-wide sharing of restricted data, (b) investigating generative model evaluation metrics, and (c) improving clinical downstream tasks. In (b), extending on common medical image synthesis assessment and reporting standards, we show Fr\'echet Inception Distance variability based on image normalisation and radiology-specific feature extraction.
Deep learning reconstruction of digital breast tomosynthesis images for accurate breast density and patient-specific radiation dose estimation
Jun 11, 2020



Abstract:The two-dimensional nature of mammography makes estimation of the overall breast density challenging, and estimation of the true patient-specific radiation dose impossible. Digital breast tomosynthesis (DBT), a pseudo-3D technique, is now commonly used in breast cancer screening and diagnostics. Still, the severely limited 3rd dimension information in DBT has not been used, until now, to estimate the true breast density or the patient-specific dose. In this study, we propose a reconstruction algorithm for DBT based on deep learning specifically optimized for these tasks. The algorithm, which we name DBToR, is based on unrolling a proximal primal-dual optimization method, where the proximal operators are replaced with convolutional neural networks and prior knowledge is included in the model. This extends previous work on a deep learning based reconstruction model by providing both the primal and the dual blocks with breast thickness information, which is available in DBT. Training and testing of the model were performed using virtual patient phantoms from two different sources. Reconstruction performance, as well as accuracy in estimation of breast density and radiation dose, was estimated, showing high accuracy (density density < +/-3%; dose < +/-20%), without bias, significantly improving on the current state-of-the-art. This work also lays the groundwork for developing a deep learning-based reconstruction algorithm for the task of image interpretation by radiologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge