Karin van Garderen
Rotterdam artery-vein segmentation (RAV) dataset
Dec 19, 2025Abstract:Purpose: To provide a diverse, high-quality dataset of color fundus images (CFIs) with detailed artery-vein (A/V) segmentation annotations, supporting the development and evaluation of machine learning algorithms for vascular analysis in ophthalmology. Methods: CFIs were sampled from the longitudinal Rotterdam Study (RS), encompassing a wide range of ages, devices, and capture conditions. Images were annotated using a custom interface that allowed graders to label arteries, veins, and unknown vessels on separate layers, starting from an initial vessel segmentation mask. Connectivity was explicitly verified and corrected using connected component visualization tools. Results: The dataset includes 1024x1024-pixel PNG images in three modalities: original RGB fundus images, contrast-enhanced versions, and RGB-encoded A/V masks. Image quality varied widely, including challenging samples typically excluded by automated quality assessment systems, but judged to contain valuable vascular information. Conclusion: This dataset offers a rich and heterogeneous source of CFIs with high-quality segmentations. It supports robust benchmarking and training of machine learning models under real-world variability in image quality and acquisition settings. Translational Relevance: By including connectivity-validated A/V masks and diverse image conditions, this dataset enables the development of clinically applicable, generalizable machine learning tools for retinal vascular analysis, potentially improving automated screening and diagnosis of systemic and ocular diseases.
VascX Models: Model Ensembles for Retinal Vascular Analysis from Color Fundus Images
Sep 24, 2024Abstract:We introduce VascX models, a comprehensive set of model ensembles for analyzing retinal vasculature from color fundus images (CFIs). Annotated CFIs were aggregated from public datasets for vessel, artery-vein, and disc segmentation; and fovea localization. Additional CFIs from the population-based Rotterdam Study were, with arteries and veins annotated by graders at pixel level. Our models achieved robust performance across devices from different vendors, varying levels of image quality levels, and diverse pathologies. Our models demonstrated superior segmentation performance compared to existing systems under a variety of conditions. Significant enhancements were observed in artery-vein and disc segmentation performance, particularly in segmentations of these structures on CFIs of intermediate quality, a common characteristic of large cohorts and clinical datasets. Our model outperformed human graders in segmenting vessels with greater precision. With VascX models we provide a robust, ready-to-use set of model ensembles and inference code aimed at simplifying the implementation and enhancing the quality of automated retinal vasculature analyses. The precise vessel parameters generated by the model can serve as starting points for the identification of disease patterns in and outside of the eye.
Deep learning-based group-wise registration for longitudinal MRI analysis in glioma
Jun 18, 2023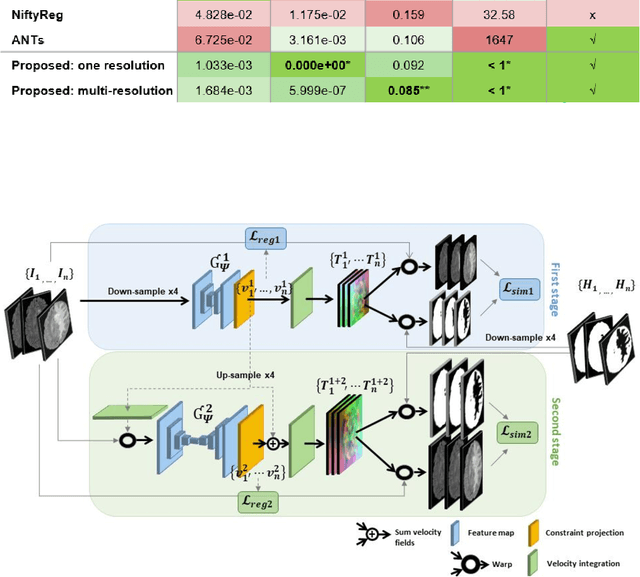
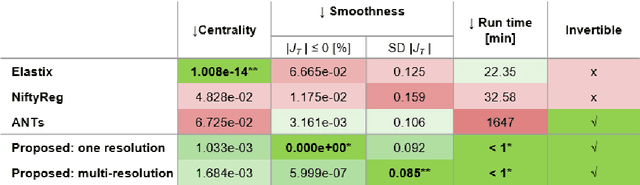
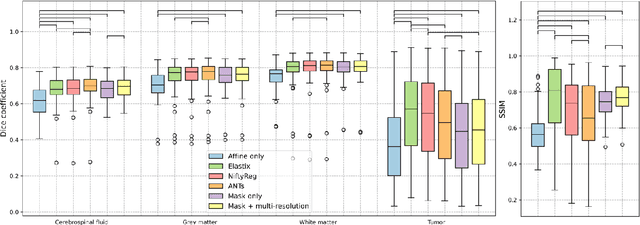
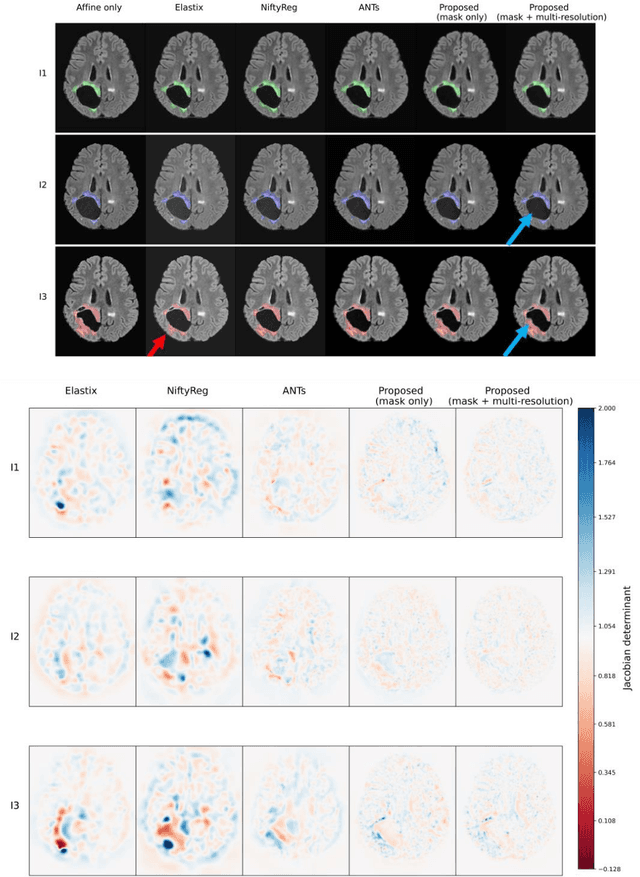
Abstract:Glioma growth may be quantified with longitudinal image registration. However, the large mass-effects and tissue changes across images pose an added challenge. Here, we propose a longitudinal, learning-based, and groupwise registration method for the accurate and unbiased registration of glioma MRI. We evaluate on a dataset from the Glioma Longitudinal AnalySiS consortium and compare it to classical registration methods. We achieve comparable Dice coefficients, with more detailed registrations, while significantly reducing the runtime to under a minute. The proposed methods may serve as an alternative to classical toolboxes, to provide further insight into glioma growth.
Towards continuous learning for glioma segmentation with elastic weight consolidation
Sep 25, 2019

Abstract:When finetuning a convolutional neural network (CNN) on data from a new domain, catastrophic forgetting will reduce performance on the original training data. Elastic Weight Consolidation (EWC) is a recent technique to prevent this, which we evaluated while training and re-training a CNN to segment glioma on two different datasets. The network was trained on the public BraTS dataset and finetuned on an in-house dataset with non-enhancing low-grade glioma. EWC was found to decrease catastrophic forgetting in this case, but was also found to restrict adaptation to the new domain.
Multi-modal segmentation with missing MR sequences using pre-trained fusion networks
Sep 25, 2019


Abstract:Missing data is a common problem in machine learning and in retrospective imaging research it is often encountered in the form of missing imaging modalities. We propose to take into account missing modalities in the design and training of neural networks, to ensure that they are capable of providing the best possible prediction even when multiple images are not available. The proposed network combines three modifications to the standard 3D UNet architecture: a training scheme with dropout of modalities, a multi-pathway architecture with fusion layer in the final stage, and the separate pre-training of these pathways. These modifications are evaluated incrementally in terms of performance on full and missing data, using the BraTS multi-modal segmentation challenge. The final model shows significant improvement with respect to the state of the art on missing data and requires less memory during training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge