Carel B. Hoyng
Uncertainty-Aware Multiple-Instance Learning for Reliable Classification: Application to Optical Coherence Tomography
Feb 06, 2023



Abstract:Deep learning classification models for medical image analysis often perform well on data from scanners that were used during training. However, when these models are applied to data from different vendors, their performance tends to drop substantially. Artifacts that only occur within scans from specific scanners are major causes of this poor generalizability. We aimed to improve the reliability of deep learning classification models by proposing Uncertainty-Based Instance eXclusion (UBIX). This technique, based on multiple-instance learning, reduces the effect of corrupted instances on the bag-classification by seamlessly integrating out-of-distribution (OOD) instance detection during inference. Although UBIX is generally applicable to different medical images and diverse classification tasks, we focused on staging of age-related macular degeneration in optical coherence tomography. After being trained using images from one vendor, UBIX showed a reliable behavior, with a slight decrease in performance (a decrease of the quadratic weighted kappa ($\kappa_w$) from 0.861 to 0.708), when applied to images from different vendors containing artifacts; while a state-of-the-art 3D neural network suffered from a significant detriment of performance ($\kappa_w$ from 0.852 to 0.084) on the same test set. We showed that instances with unseen artifacts can be identified with OOD detection and their contribution to the bag-level predictions can be reduced, improving reliability without the need for retraining on new data. This potentially increases the applicability of artificial intelligence models to data from other scanners than the ones for which they were developed.
A deep learning model for segmentation of geographic atrophy to study its long-term natural history
Aug 15, 2019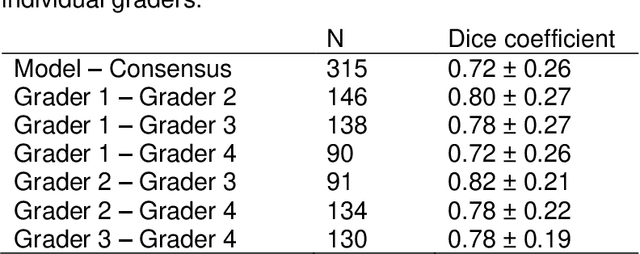
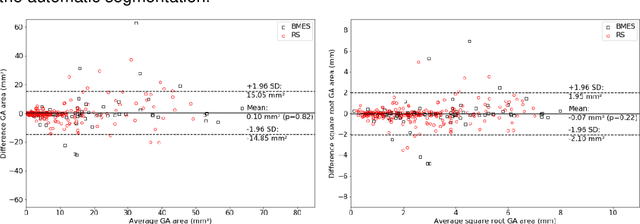
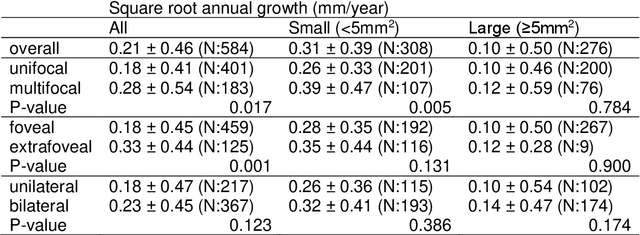
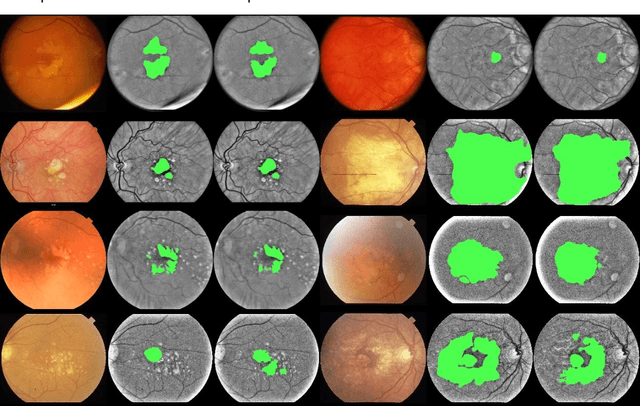
Abstract:Purpose: To develop and validate a deep learning model for automatic segmentation of geographic atrophy (GA) in color fundus images (CFIs) and its application to study growth rate of GA. Participants: 409 CFIs of 238 eyes with GA from the Rotterdam Study (RS) and the Blue Mountain Eye Study (BMES) for model development, and 5,379 CFIs of 625 eyes from the Age-Related Eye Disease Study (AREDS) for analysis of GA growth rate. Methods: A deep learning model based on an ensemble of encoder-decoder architectures was implemented and optimized for the segmentation of GA in CFIs. Four experienced graders delineated GA in CFIs from RS and BMES. These manual delineations were used to evaluate the segmentation model using 5-fold cross-validation. The model was further applied to CFIs from the AREDS to study the growth rate of GA. Linear regression analysis was used to study associations between structural biomarkers at baseline and GA growth rate. A general estimate of the progression of GA area over time was made by combining growth rates of all eyes with GA from the AREDS set. Results: The model obtained an average Dice coefficient of 0.72 $\pm$ 0.26 on the BMES and RS. An intraclass correlation coefficient of 0.83 was reached between the automatically estimated GA area and the graders' consensus measures. Eight automatically calculated structural biomarkers (area, filled area, convex area, convex solidity, eccentricity, roundness, foveal involvement and perimeter) were significantly associated with growth rate. Combining all growth rates indicated that GA area grows quadratically up to an area of around 12 mm$^{2}$, after which growth rate stabilizes or decreases. Conclusion: The presented deep learning model allowed for fully automatic and robust segmentation of GA in CFIs. These segmentations can be used to extract structural characteristics of GA that predict its growth rate.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge