Hoel Kervadec
Zero-shot capability of SAM-family models for bone segmentation in CT scans
Nov 13, 2024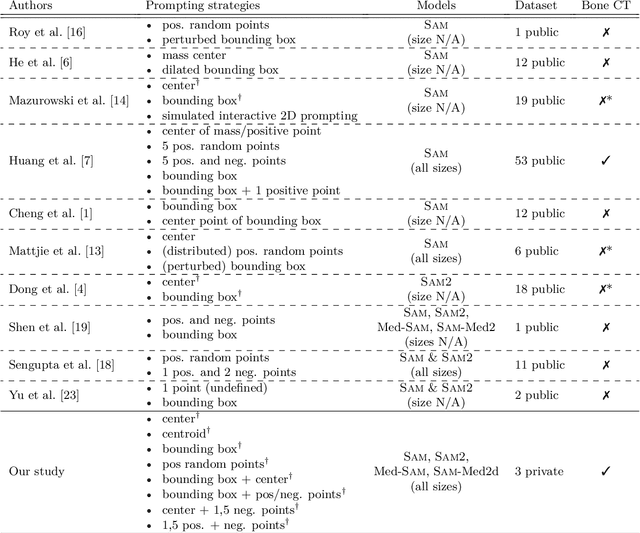

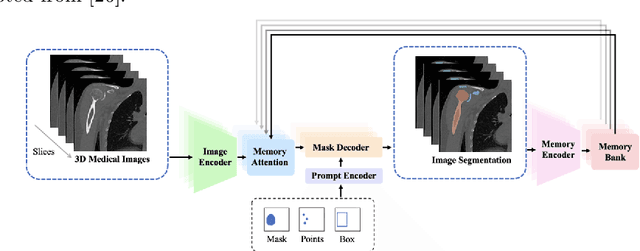
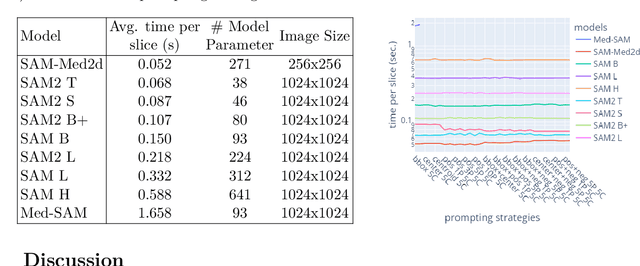
Abstract:The Segment Anything Model (SAM) and similar models build a family of promptable foundation models (FMs) for image and video segmentation. The object of interest is identified using prompts, such as bounding boxes or points. With these FMs becoming part of medical image segmentation, extensive evaluation studies are required to assess their strengths and weaknesses in clinical setting. Since the performance is highly dependent on the chosen prompting strategy, it is important to investigate different prompting techniques to define optimal guidelines that ensure effective use in medical image segmentation. Currently, no dedicated evaluation studies exist specifically for bone segmentation in CT scans, leaving a gap in understanding the performance for this task. Thus, we use non-iterative, ``optimal'' prompting strategies composed of bounding box, points and combinations to test the zero-shot capability of SAM-family models for bone CT segmentation on three different skeletal regions. Our results show that the best settings depend on the model type and size, dataset characteristics and objective to optimize. Overall, SAM and SAM2 prompted with a bounding box in combination with the center point for all the components of an object yield the best results across all tested settings. As the results depend on multiple factors, we provide a guideline for informed decision-making in 2D prompting with non-interactive, ''optimal'' prompts.
Leveraging point annotations in segmentation learning with boundary loss
Nov 06, 2023Abstract:This paper investigates the combination of intensity-based distance maps with boundary loss for point-supervised semantic segmentation. By design the boundary loss imposes a stronger penalty on the false positives the farther away from the object they occur. Hence it is intuitively inappropriate for weak supervision, where the ground truth label may be much smaller than the actual object and a certain amount of false positives (w.r.t. the weak ground truth) is actually desirable. Using intensity-aware distances instead may alleviate this drawback, allowing for a certain amount of false positives without a significant increase to the training loss. The motivation for applying the boundary loss directly under weak supervision lies in its great success for fully supervised segmentation tasks, but also in not requiring extra priors or outside information that is usually required -- in some form -- with existing weakly supervised methods in the literature. This formulation also remains potentially more attractive than existing CRF-based regularizers, due to its simplicity and computational efficiency. We perform experiments on two multi-class datasets; ACDC (heart segmentation) and POEM (whole-body abdominal organ segmentation). Preliminary results are encouraging and show that this supervision strategy has great potential. On ACDC it outperforms the CRF-loss based approach, and on POEM data it performs on par with it. The code for all our experiments is openly available.
On the dice loss gradient and the ways to mimic it
Apr 09, 2023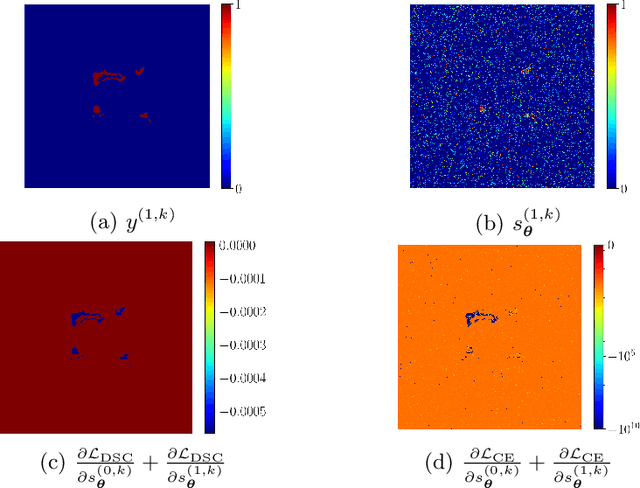
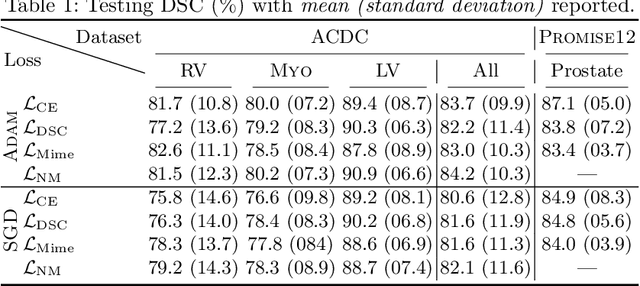
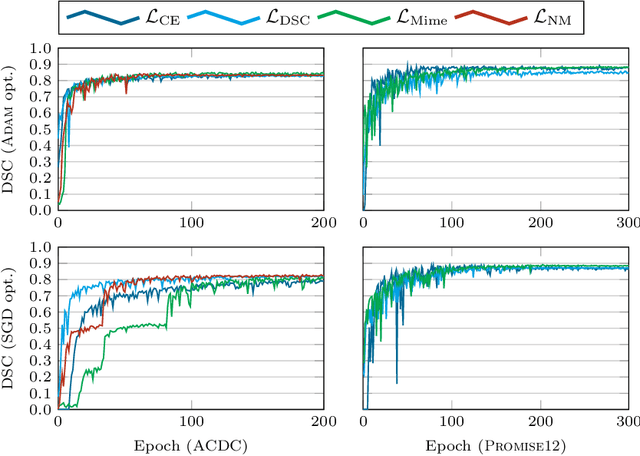
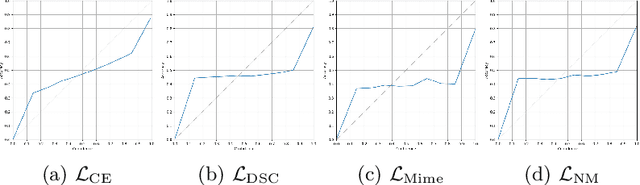
Abstract:In the past few years, in the context of fully-supervised semantic segmentation, several losses -- such as cross-entropy and dice -- have emerged as de facto standards to supervise neural networks. The Dice loss is an interesting case, as it comes from the relaxation of the popular Dice coefficient; one of the main evaluation metric in medical imaging applications. In this paper, we first study theoretically the gradient of the dice loss, showing that concretely it is a weighted negative of the ground truth, with a very small dynamic range. This enables us, in the second part of this paper, to mimic the supervision of the dice loss, through a simple element-wise multiplication of the network output with a negative of the ground truth. This rather surprising result sheds light on the practical supervision performed by the dice loss during gradient descent. This can help the practitioner to understand and interpret results while guiding researchers when designing new losses.
Source-Free Domain Adaptation for Image Segmentation
Aug 06, 2021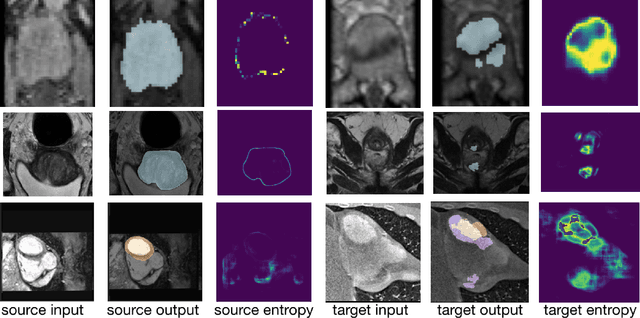
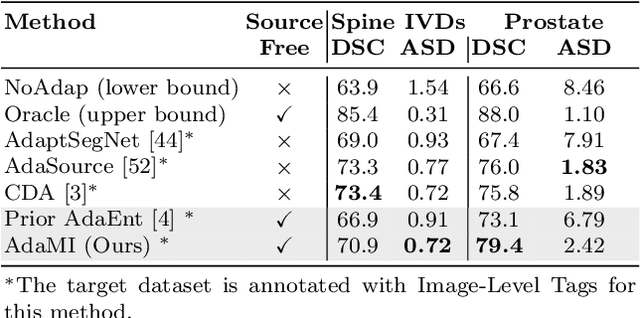
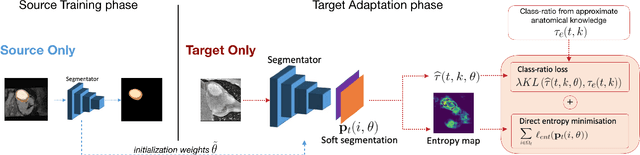
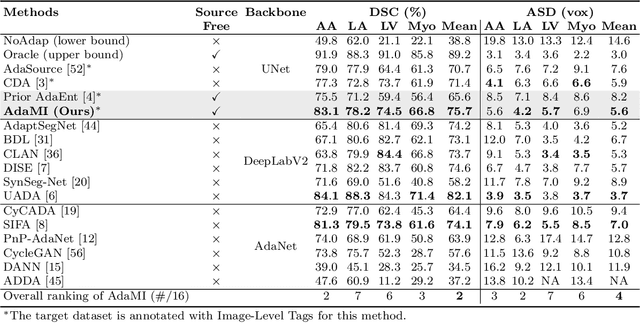
Abstract:Domain adaptation (DA) has drawn high interest for its capacity to adapt a model trained on labeled source data to perform well on unlabeled or weakly labeled target data from a different domain. Most common DA techniques require concurrent access to the input images of both the source and target domains. However, in practice, privacy concerns often impede the availability of source images in the adaptation phase. This is a very frequent DA scenario in medical imaging, where, for instance, the source and target images could come from different clinical sites. We introduce a source-free domain adaptation for image segmentation. Our formulation is based on minimizing a label-free entropy loss defined over target-domain data, which we further guide with a domain-invariant prior on the segmentation regions. Many priors can be derived from anatomical information. Here, a class ratio prior is estimated from anatomical knowledge and integrated in the form of a Kullback Leibler (KL) divergence in our overall loss function. Furthermore, we motivate our overall loss with an interesting link to maximizing the mutual information between the target images and their label predictions. We show the effectiveness of our prior aware entropy minimization in a variety of domain-adaptation scenarios, with different modalities and applications, including spine, prostate, and cardiac segmentation. Our method yields comparable results to several state of the art adaptation techniques, despite having access to much less information, as the source images are entirely absent in our adaptation phase. Our straightforward adaptation strategy uses only one network, contrary to popular adversarial techniques, which are not applicable to a source-free DA setting. Our framework can be readily used in a breadth of segmentation problems, and our code is publicly available: https://github.com/mathilde-b/SFDA
Beyond pixel-wise supervision for segmentation: A few global shape descriptors might be surprisingly good!
May 03, 2021

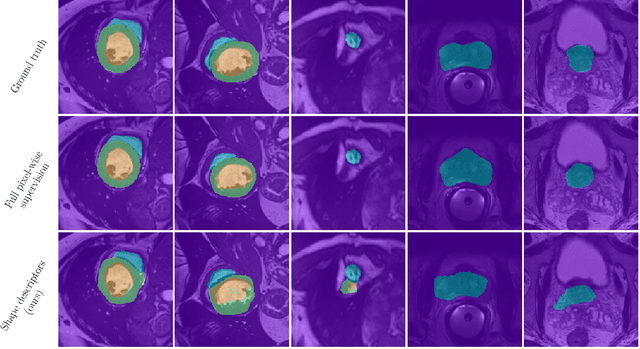

Abstract:Standard losses for training deep segmentation networks could be seen as individual classifications of pixels, instead of supervising the global shape of the predicted segmentations. While effective, they require exact knowledge of the label of each pixel in an image. This study investigates how effective global geometric shape descriptors could be, when used on their own as segmentation losses for training deep networks. Not only interesting theoretically, there exist deeper motivations to posing segmentation problems as a reconstruction of shape descriptors: Annotations to obtain approximations of low-order shape moments could be much less cumbersome than their full-mask counterparts, and anatomical priors could be readily encoded into invariant shape descriptions, which might alleviate the annotation burden. Also, and most importantly, we hypothesize that, given a task, certain shape descriptions might be invariant across image acquisition protocols/modalities and subject populations, which might open interesting research avenues for generalization in medical image segmentation. We introduce and formulate a few shape descriptors in the context of deep segmentation, and evaluate their potential as standalone losses on two different challenging tasks. Inspired by recent works in constrained optimization for deep networks, we propose a way to use those descriptors to supervise segmentation, without any pixel-level label. Very surprisingly, as little as 4 descriptors values per class can approach the performance of a segmentation mask with 65k individual discrete labels. We also found that shape descriptors can be a valid way to encode anatomical priors about the task, enabling to leverage expert knowledge without additional annotations. Our implementation is publicly available and can be easily extended to other tasks and descriptors: https://github.com/hkervadec/shape_descriptors
Few-Shot Segmentation Without Meta-Learning: A Good Transductive Inference Is All You Need?
Dec 11, 2020
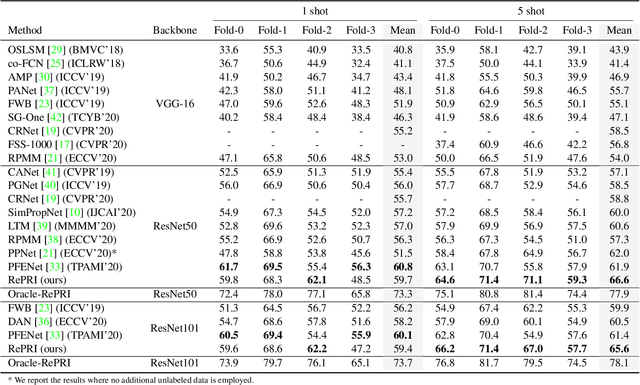
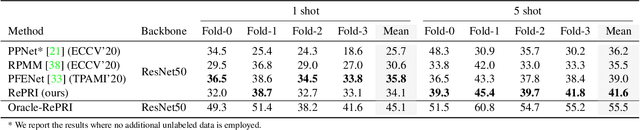
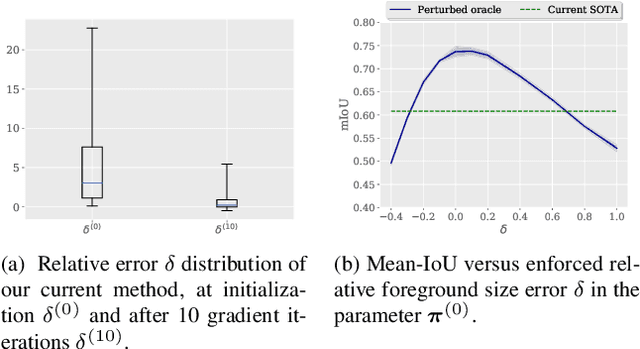
Abstract:Few-shot segmentation has recently attracted substantial interest, with the popular meta-learning paradigm widely dominating the literature. We show that the way inference is performed for a given few-shot segmentation task has a substantial effect on performances, an aspect that has been overlooked in the literature. We introduce a transductive inference, which leverages the statistics of the unlabeled pixels of a task by optimizing a new loss containing three complementary terms: (i) a standard cross-entropy on the labeled pixels; (ii) the entropy of posteriors on the unlabeled query pixels; and (iii) a global KL-divergence regularizer based on the proportion of the predicted foreground region. Our inference uses a simple linear classifier of the extracted features, has a computational load comparable to inductive inference and can be used on top of any base training. Using standard cross-entropy training on the base classes, our inference yields highly competitive performances on well-known few-shot segmentation benchmarks. On PASCAL-5i, it brings about 5% improvement over the best performing state-of-the-art method in the 5-shot scenario, while being on par in the 1-shot setting. Even more surprisingly, this gap widens as the number of support samples increases, reaching up to 6% in the 10-shot scenario. Furthermore, we introduce a more realistic setting with domain shift, where the base and novel classes are drawn from different datasets. In this setting, we found that our method achieves the best performances.
Source-Relaxed Domain Adaptation for Image Segmentation
May 07, 2020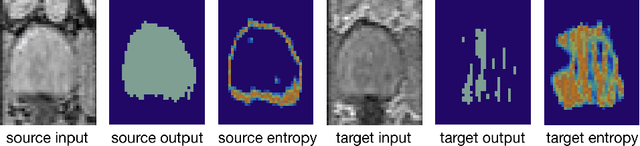
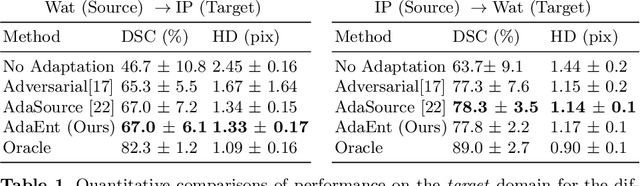
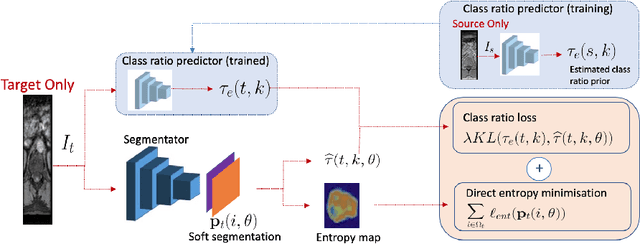
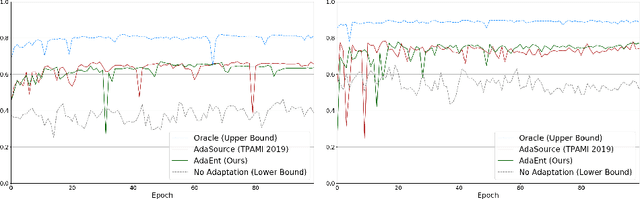
Abstract:Domain adaptation (DA) has drawn high interests for its capacity to adapt a model trained on labeled source data to perform well on unlabeled or weakly labeled target data from a different domain. Most common DA techniques require the concurrent access to the input images of both the source and target domains. However, in practice, it is common that the source images are not available in the adaptation phase. This is a very frequent DA scenario in medical imaging, for instance, when the source and target images come from different clinical sites. We propose a novel formulation for adapting segmentation networks, which relaxes such a constraint. Our formulation is based on minimizing a label-free entropy loss defined over target-domain data, which we further guide with a domain invariant prior on the segmentation regions. Many priors can be used, derived from anatomical information. Here, a class-ratio prior is learned via an auxiliary network and integrated in the form of a Kullback-Leibler (KL) divergence in our overall loss function. We show the effectiveness of our prior-aware entropy minimization in adapting spine segmentation across different MRI modalities. Our method yields comparable results to several state-of-the-art adaptation techniques, even though is has access to less information, the source images being absent in the adaptation phase. Our straight-forward adaptation strategy only uses one network, contrary to popular adversarial techniques, which cannot perform without the presence of the source images. Our framework can be readily used with various priors and segmentation problems.
Bounding boxes for weakly supervised segmentation: Global constraints get close to full supervision
Apr 14, 2020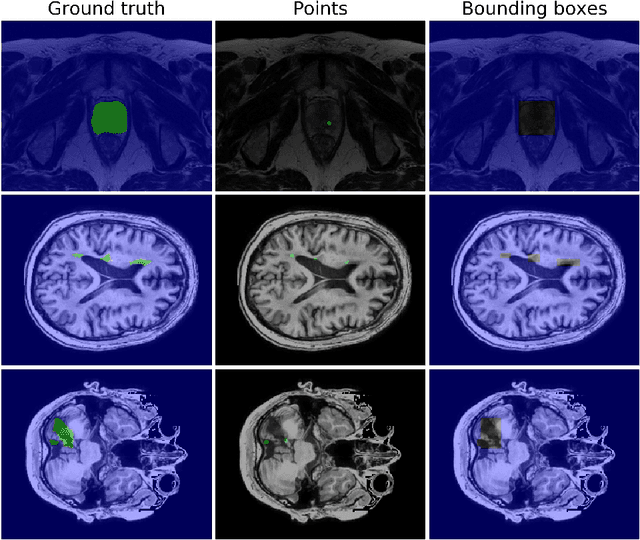
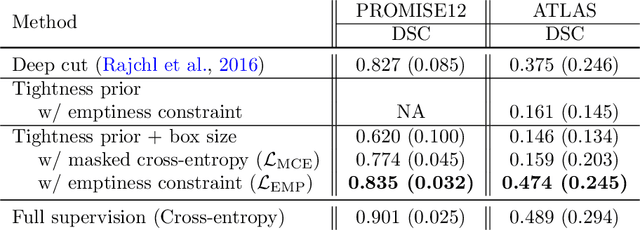
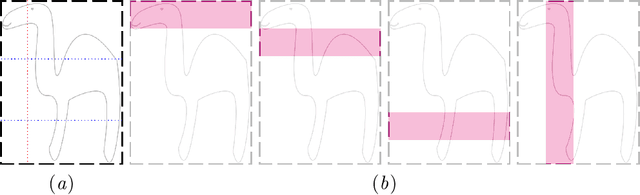

Abstract:We propose a novel weakly supervised learning segmentation based on several global constraints derived from box annotations. Particularly, we leverage a classical tightness prior to a deep learning setting via imposing a set of constraints on the network outputs. Such a powerful topological prior prevents solutions from excessive shrinking by enforcing any horizontal or vertical line within the bounding box to contain, at least, one pixel of the foreground region. Furthermore, we integrate our deep tightness prior with a global background emptiness constraint, guiding training with information outside the bounding box. We demonstrate experimentally that such a global constraint is much more powerful than standard cross-entropy for the background class. Our optimization problem is challenging as it takes the form of a large set of inequality constraints on the outputs of deep networks. We solve it with sequence of unconstrained losses based on a recent powerful extension of the log-barrier method, which is well-known in the context of interior-point methods. This accommodates standard stochastic gradient descent (SGD) for training deep networks, while avoiding computationally expensive and unstable Lagrangian dual steps and projections. Extensive experiments over two different public data sets and applications (prostate and brain lesions) demonstrate that the synergy between our global tightness and emptiness priors yield very competitive performances, approaching full supervision and outperforming significantly DeepCut. Furthermore, our approach removes the need for computationally expensive proposal generation. Our code is shared anonymously.
Discretely-constrained deep network for weakly supervised segmentation
Aug 15, 2019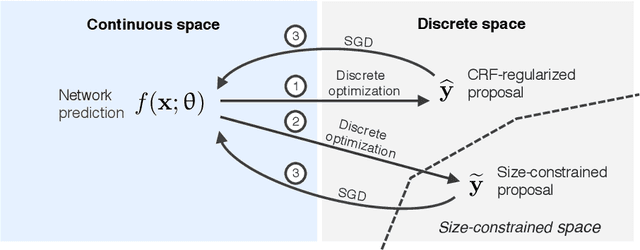
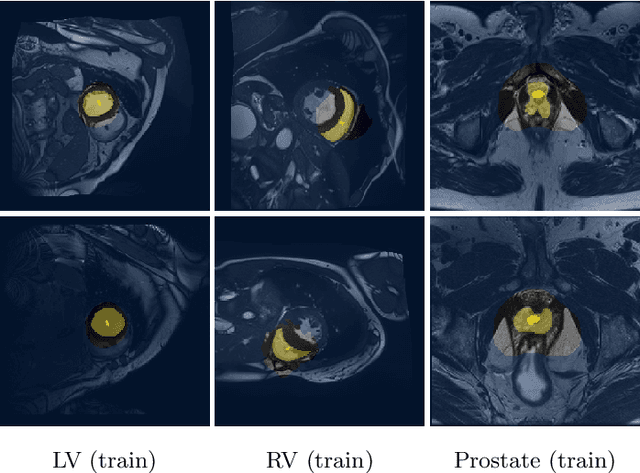
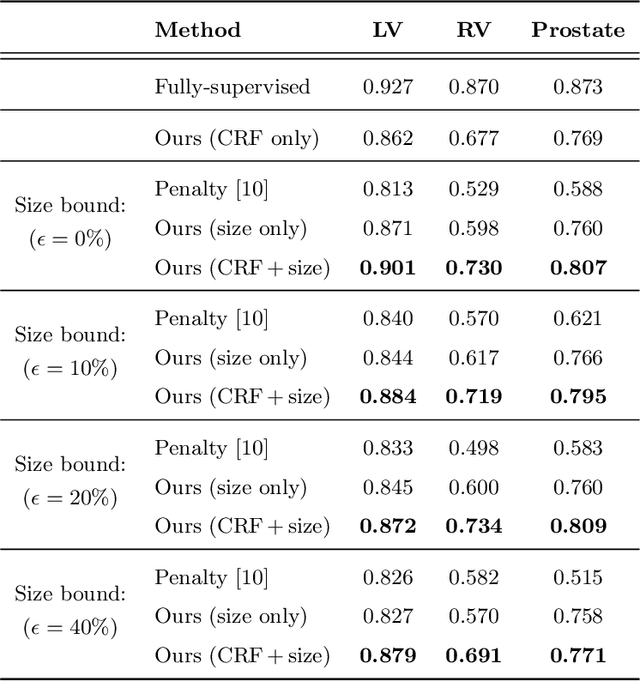
Abstract:An efficient strategy for weakly-supervised segmentation is to impose constraints or regularization priors on target regions. Recent efforts have focused on incorporating such constraints in the training of convolutional neural networks (CNN), however this has so far been done within a continuous optimization framework. Yet, various segmentation constraints and regularization can be modeled and optimized more efficiently in a discrete formulation. This paper proposes a method, based on the alternating direction method of multipliers (ADMM) algorithm, to train a CNN with discrete constraints and regularization priors. This method is applied to the segmentation of medical images with weak annotations, where both size constraints and boundary length regularization are enforced. Experiments on a benchmark cardiac segmentation dataset show our method to yield a performance near to full supervision.
Constrained domain adaptation for segmentation
Aug 08, 2019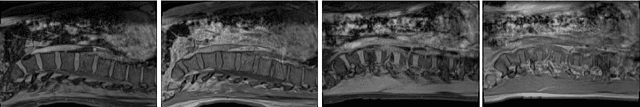
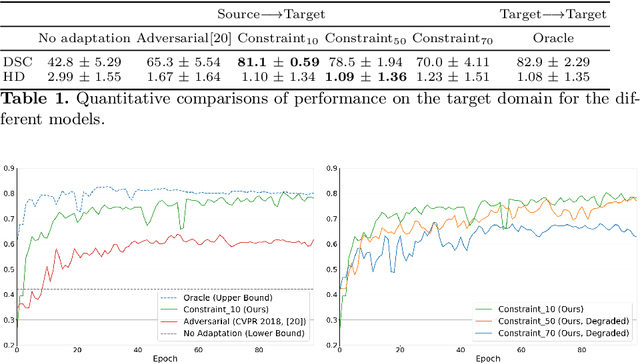
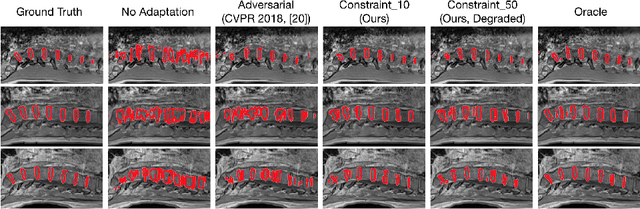
Abstract:We propose to adapt segmentation networks with a constrained formulation, which embeds domain-invariant prior knowledge about the segmentation regions. Such knowledge may take the form of simple anatomical information, e.g., structure size or shape, estimated from source samples or known a priori. Our method imposes domain-invariant inequality constraints on the network outputs of unlabeled target samples. It implicitly matches prediction statistics between target and source domains with permitted uncertainty of prior knowledge. We address our constrained problem with a differentiable penalty, fully suited for standard stochastic gradient descent approaches, removing the need for computationally expensive Lagrangian optimization with dual projections. Unlike current two-step adversarial training, our formulation is based on a single loss in a single network, which simplifies adaptation by avoiding extra adversarial steps, while improving convergence and quality of training. The comparison of our approach with state-of-the-art adversarial methods reveals substantially better performance on the challenging task of adapting spine segmentation across different MRI modalities. Our results also show a robustness to imprecision of size priors, approaching the accuracy of a fully supervised model trained directly in a target domain.Our method can be readily used for various constraints and segmentation problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge