Herve Lombaert
Spectral State Space Model for Rotation-Invariant~Visual~Representation~Learning
Mar 09, 2025Abstract:State Space Models (SSMs) have recently emerged as an alternative to Vision Transformers (ViTs) due to their unique ability of modeling global relationships with linear complexity. SSMs are specifically designed to capture spatially proximate relationships of image patches. However, they fail to identify relationships between conceptually related yet not adjacent patches. This limitation arises from the non-causal nature of image data, which lacks inherent directional relationships. Additionally, current vision-based SSMs are highly sensitive to transformations such as rotation. Their predefined scanning directions depend on the original image orientation, which can cause the model to produce inconsistent patch-processing sequences after rotation. To address these limitations, we introduce Spectral VMamba, a novel approach that effectively captures the global structure within an image by leveraging spectral information derived from the graph Laplacian of image patches. Through spectral decomposition, our approach encodes patch relationships independently of image orientation, achieving rotation invariance with the aid of our Rotational Feature Normalizer (RFN) module. Our experiments on classification tasks show that Spectral VMamba outperforms the leading SSM models in vision, such as VMamba, while maintaining invariance to rotations and a providing a similar runtime efficiency.
Sparse Bayesian Networks: Efficient Uncertainty Quantification in Medical Image Analysis
Jun 11, 2024Abstract:Efficiently quantifying predictive uncertainty in medical images remains a challenge. While Bayesian neural networks (BNN) offer predictive uncertainty, they require substantial computational resources to train. Although Bayesian approximations such as ensembles have shown promise, they still suffer from high training and inference costs. Existing approaches mainly address the costs of BNN inference post-training, with little focus on improving training efficiency and reducing parameter complexity. This study introduces a training procedure for a sparse (partial) Bayesian network. Our method selectively assigns a subset of parameters as Bayesian by assessing their deterministic saliency through gradient sensitivity analysis. The resulting network combines deterministic and Bayesian parameters, exploiting the advantages of both representations to achieve high task-specific performance and minimize predictive uncertainty. Demonstrated on multi-label ChestMNIST for classification and ISIC, LIDC-IDRI for segmentation, our approach achieves competitive performance and predictive uncertainty estimation by reducing Bayesian parameters by over 95\%, significantly reducing computational expenses compared to fully Bayesian and ensemble methods.
Anatomically-aware Uncertainty for Semi-supervised Image Segmentation
Oct 24, 2023Abstract:Semi-supervised learning relaxes the need of large pixel-wise labeled datasets for image segmentation by leveraging unlabeled data. A prominent way to exploit unlabeled data is to regularize model predictions. Since the predictions of unlabeled data can be unreliable, uncertainty-aware schemes are typically employed to gradually learn from meaningful and reliable predictions. Uncertainty estimation methods, however, rely on multiple inferences from the model predictions that must be computed for each training step, which is computationally expensive. Moreover, these uncertainty maps capture pixel-wise disparities and do not consider global information. This work proposes a novel method to estimate segmentation uncertainty by leveraging global information from the segmentation masks. More precisely, an anatomically-aware representation is first learnt to model the available segmentation masks. The learnt representation thereupon maps the prediction of a new segmentation into an anatomically-plausible segmentation. The deviation from the plausible segmentation aids in estimating the underlying pixel-level uncertainty in order to further guide the segmentation network. The proposed method consequently estimates the uncertainty using a single inference from our representation, thereby reducing the total computation. We evaluate our method on two publicly available segmentation datasets of left atria in cardiac MRIs and of multiple organs in abdominal CTs. Our anatomically-aware method improves the segmentation accuracy over the state-of-the-art semi-supervised methods in terms of two commonly used evaluation metrics.
Attention-based Dynamic Subspace Learners for Medical Image Analysis
Jun 18, 2022



Abstract:Learning similarity is a key aspect in medical image analysis, particularly in recommendation systems or in uncovering the interpretation of anatomical data in images. Most existing methods learn such similarities in the embedding space over image sets using a single metric learner. Images, however, have a variety of object attributes such as color, shape, or artifacts. Encoding such attributes using a single metric learner is inadequate and may fail to generalize. Instead, multiple learners could focus on separate aspects of these attributes in subspaces of an overarching embedding. This, however, implies the number of learners to be found empirically for each new dataset. This work, Dynamic Subspace Learners, proposes to dynamically exploit multiple learners by removing the need of knowing apriori the number of learners and aggregating new subspace learners during training. Furthermore, the visual interpretability of such subspace learning is enforced by integrating an attention module into our method. This integrated attention mechanism provides a visual insight of discriminative image features that contribute to the clustering of image sets and a visual explanation of the embedding features. The benefits of our attention-based dynamic subspace learners are evaluated in the application of image clustering, image retrieval, and weakly supervised segmentation. Our method achieves competitive results with the performances of multiple learners baselines and significantly outperforms the classification network in terms of clustering and retrieval scores on three different public benchmark datasets. Moreover, our attention maps offer a proxy-labels, which improves the segmentation accuracy up to 15% in Dice scores when compared to state-of-the-art interpretation techniques.
Leveraging Labeling Representations in Uncertainty-based Semi-supervised Segmentation
Mar 10, 2022



Abstract:Semi-supervised segmentation tackles the scarcity of annotations by leveraging unlabeled data with a small amount of labeled data. A prominent way to utilize the unlabeled data is by consistency training which commonly uses a teacher-student network, where a teacher guides a student segmentation. The predictions of unlabeled data are not reliable, therefore, uncertainty-aware methods have been proposed to gradually learn from meaningful and reliable predictions. Uncertainty estimation, however, relies on multiple inferences from model predictions that need to be computed for each training step, which is computationally expensive. This work proposes a novel method to estimate the pixel-level uncertainty by leveraging the labeling representation of segmentation masks. On the one hand, a labeling representation is learnt to represent the available segmentation masks. The learnt labeling representation is used to map the prediction of the segmentation into a set of plausible masks. Such a reconstructed segmentation mask aids in estimating the pixel-level uncertainty guiding the segmentation network. The proposed method estimates the uncertainty with a single inference from the labeling representation, thereby reducing the total computation. We evaluate our method on the 3D segmentation of left atrium in MRI, and we show that our uncertainty estimates from our labeling representation improve the segmentation accuracy over state-of-the-art methods.
Medial Spectral Coordinates for 3D Shape Analysis
Nov 30, 2021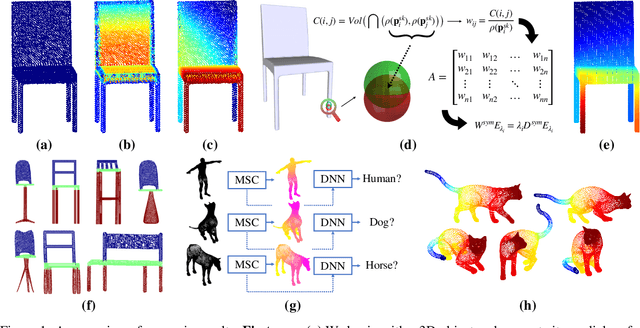
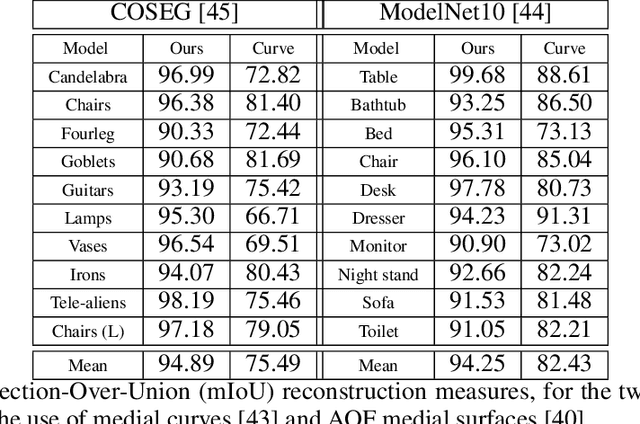
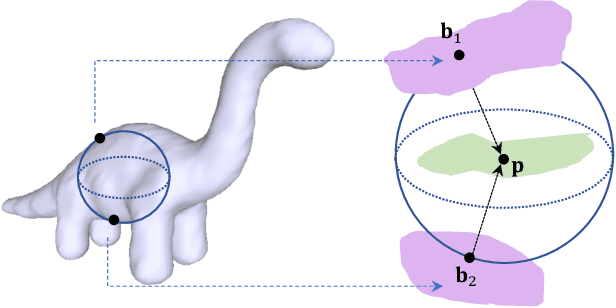
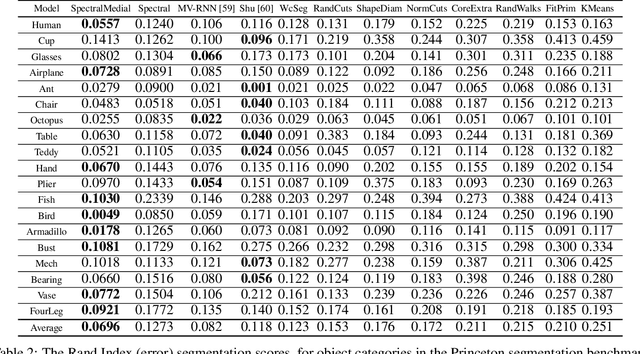
Abstract:In recent years there has been a resurgence of interest in our community in the shape analysis of 3D objects represented by surface meshes, their voxelized interiors, or surface point clouds. In part, this interest has been stimulated by the increased availability of RGBD cameras, and by applications of computer vision to autonomous driving, medical imaging, and robotics. In these settings, spectral coordinates have shown promise for shape representation due to their ability to incorporate both local and global shape properties in a manner that is qualitatively invariant to isometric transformations. Yet, surprisingly, such coordinates have thus far typically considered only local surface positional or derivative information. In the present article, we propose to equip spectral coordinates with medial (object width) information, so as to enrich them. The key idea is to couple surface points that share a medial ball, via the weights of the adjacency matrix. We develop a spectral feature using this idea, and the algorithms to compute it. The incorporation of object width and medial coupling has direct benefits, as illustrated by our experiments on object classification, object part segmentation, and surface point correspondence.
Realistic Image Normalization for Multi-Domain Segmentation
Oct 02, 2020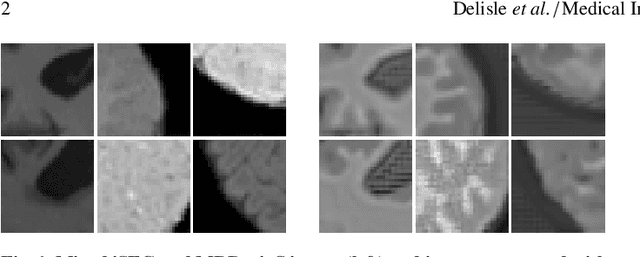
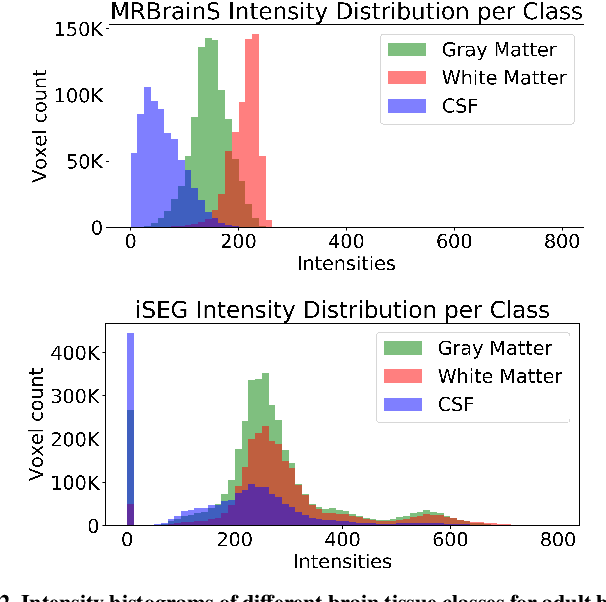
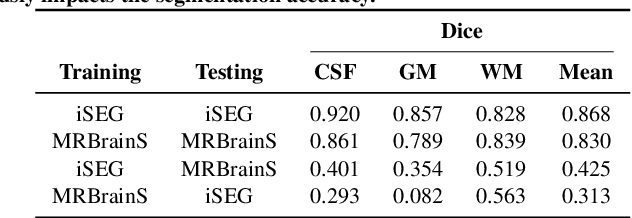
Abstract:Image normalization is a building block in medical image analysis. Conventional approaches are customarily utilized on a per-dataset basis. This strategy, however, prevents the current normalization algorithms from fully exploiting the complex joint information available across multiple datasets. Consequently, ignoring such joint information has a direct impact on the performance of segmentation algorithms. This paper proposes to revisit the conventional image normalization approach by instead learning a common normalizing function across multiple datasets. Jointly normalizing multiple datasets is shown to yield consistent normalized images as well as an improved image segmentation. To do so, a fully automated adversarial and task-driven normalization approach is employed as it facilitates the training of realistic and interpretable images while keeping performance on-par with the state-of-the-art. The adversarial training of our network aims at finding the optimal transfer function to improve both the segmentation accuracy and the generation of realistic images. We evaluated the performance of our normalizer on both infant and adult brains images from the iSEG, MRBrainS and ABIDE datasets. Results reveal the potential of our normalization approach for segmentation, with Dice improvements of up to 57.5% over our baseline. Our method can also enhance data availability by increasing the number of samples available when learning from multiple imaging domains.
Medical Imaging with Deep Learning: MIDL 2020 -- Short Paper Track
Jun 29, 2020Abstract:This compendium gathers all the accepted extended abstracts from the Third International Conference on Medical Imaging with Deep Learning (MIDL 2020), held in Montreal, Canada, 6-9 July 2020. Note that only accepted extended abstracts are listed here, the Proceedings of the MIDL 2020 Full Paper Track are published in the Proceedings of Machine Learning Research (PMLR).
Source-Relaxed Domain Adaptation for Image Segmentation
May 07, 2020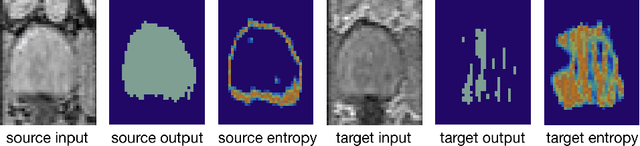
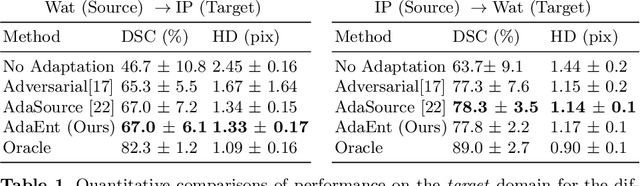
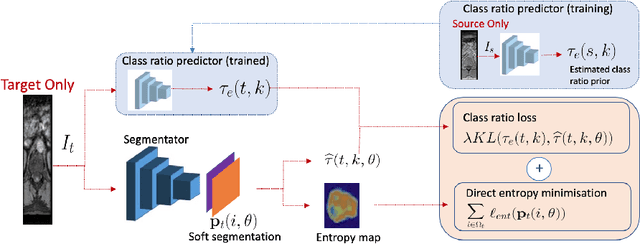
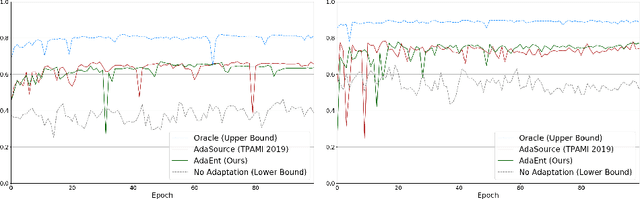
Abstract:Domain adaptation (DA) has drawn high interests for its capacity to adapt a model trained on labeled source data to perform well on unlabeled or weakly labeled target data from a different domain. Most common DA techniques require the concurrent access to the input images of both the source and target domains. However, in practice, it is common that the source images are not available in the adaptation phase. This is a very frequent DA scenario in medical imaging, for instance, when the source and target images come from different clinical sites. We propose a novel formulation for adapting segmentation networks, which relaxes such a constraint. Our formulation is based on minimizing a label-free entropy loss defined over target-domain data, which we further guide with a domain invariant prior on the segmentation regions. Many priors can be used, derived from anatomical information. Here, a class-ratio prior is learned via an auxiliary network and integrated in the form of a Kullback-Leibler (KL) divergence in our overall loss function. We show the effectiveness of our prior-aware entropy minimization in adapting spine segmentation across different MRI modalities. Our method yields comparable results to several state-of-the-art adaptation techniques, even though is has access to less information, the source images being absent in the adaptation phase. Our straight-forward adaptation strategy only uses one network, contrary to popular adversarial techniques, which cannot perform without the presence of the source images. Our framework can be readily used with various priors and segmentation problems.
Manifold-Aware CycleGAN for High Resolution Structural-to-DTI Synthesis
Apr 08, 2020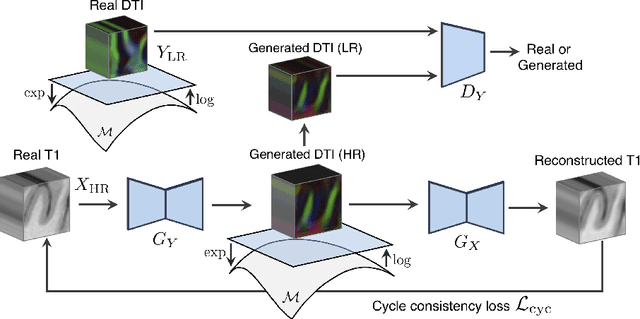
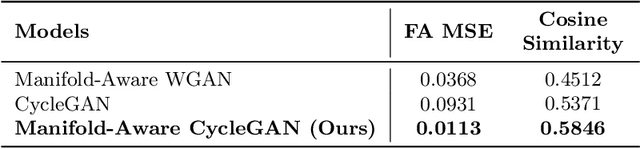
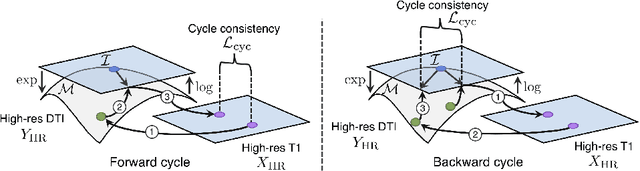
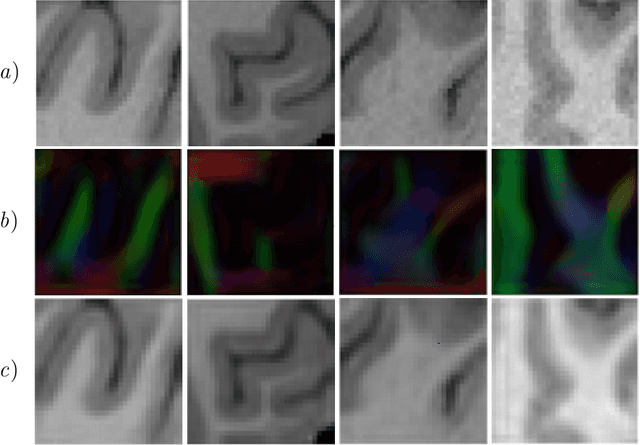
Abstract:Unpaired image-to-image translation has been applied successfully to natural images but has received very little attention for manifold-valued data such as in diffusion tensor imaging (DTI). The non-Euclidean nature of DTI prevents current generative adversarial networks (GANs) from generating plausible images and has mostly limited their application to diffusion MRI scalar maps, such as fractional anisotropy (FA) or mean diffusivity (MD). Even if these scalar maps are clinically useful, they mostly ignore fiber orientations and have, therefore, limited applications for analyzing brain fibers, for instance, impairing fiber tractography. Here, we propose a manifold-aware CycleGAN that learns the generation of high resolution DTI from unpaired T1w images. We formulate the objective as a Wasserstein distance minimization problem of data distributions on a Riemannian manifold of symmetric positive definite 3x3 matrices SPD(3), using adversarial and cycle-consistency losses. To ensure that the generated diffusion tensors lie on the SPD(3) manifold, we exploit the theoretical properties of the exponential and logarithm maps. We demonstrate that, unlike standard GANs, our method is able to generate realistic high resolution DTI that can be used to compute diffusion-based metrics and run fiber tractography algorithms. To evaluate our model's performance, we compute the cosine similarity between the generated tensors principal orientation and their ground truth orientation and the mean squared error (MSE) of their derived FA values. We demonstrate that our method produces up to 8 times better FA MSE than a standard CycleGAN and 30% better cosine similarity than a manifold-aware Wasserstein GAN while synthesizing sharp high resolution DTI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge