Samuel Kadoury
Polytechnique Montreal, Canada, CHU Sainte-Justine Research Center, Montreal, Canada
Sparse Bayesian Networks: Efficient Uncertainty Quantification in Medical Image Analysis
Jun 11, 2024Abstract:Efficiently quantifying predictive uncertainty in medical images remains a challenge. While Bayesian neural networks (BNN) offer predictive uncertainty, they require substantial computational resources to train. Although Bayesian approximations such as ensembles have shown promise, they still suffer from high training and inference costs. Existing approaches mainly address the costs of BNN inference post-training, with little focus on improving training efficiency and reducing parameter complexity. This study introduces a training procedure for a sparse (partial) Bayesian network. Our method selectively assigns a subset of parameters as Bayesian by assessing their deterministic saliency through gradient sensitivity analysis. The resulting network combines deterministic and Bayesian parameters, exploiting the advantages of both representations to achieve high task-specific performance and minimize predictive uncertainty. Demonstrated on multi-label ChestMNIST for classification and ISIC, LIDC-IDRI for segmentation, our approach achieves competitive performance and predictive uncertainty estimation by reducing Bayesian parameters by over 95\%, significantly reducing computational expenses compared to fully Bayesian and ensemble methods.
Generative Adversarial Networks in Ultrasound Imaging: Extending Field of View Beyond Conventional Limits
May 31, 2024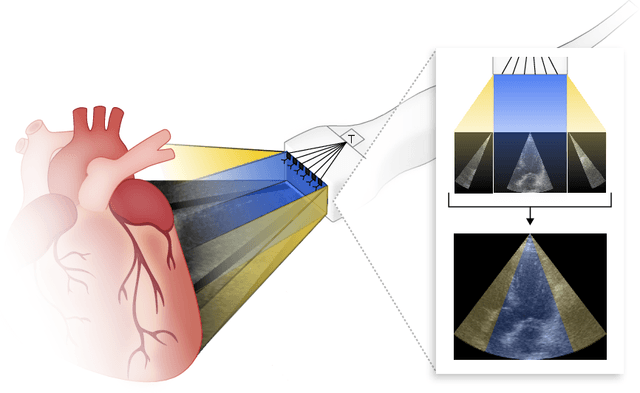
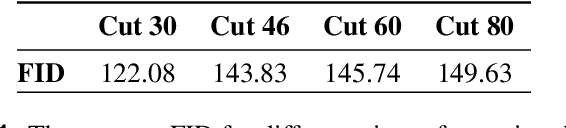
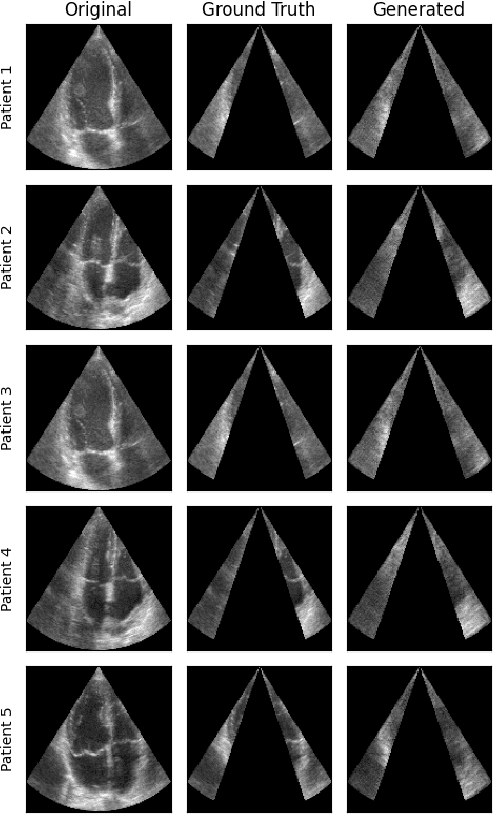
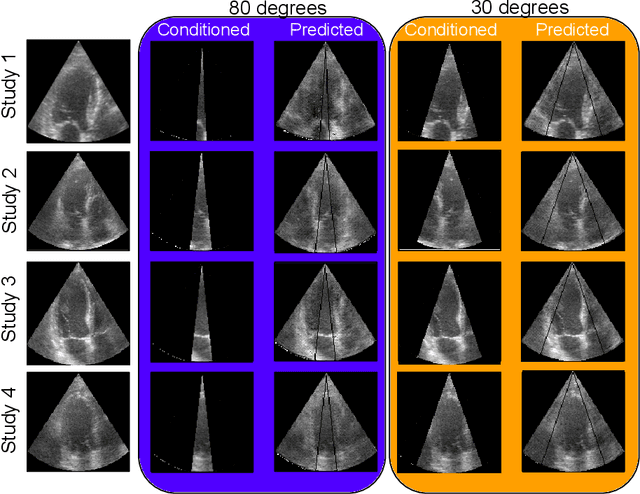
Abstract:Transthoracic Echocardiography (TTE) is a fundamental, non-invasive diagnostic tool in cardiovascular medicine, enabling detailed visualization of cardiac structures crucial for diagnosing various heart conditions. Despite its widespread use, TTE ultrasound imaging faces inherent limitations, notably the trade-off between field of view (FoV) and resolution. This paper introduces a novel application of conditional Generative Adversarial Networks (cGANs), specifically designed to extend the FoV in TTE ultrasound imaging while maintaining high resolution. Our proposed cGAN architecture, termed echoGAN, demonstrates the capability to generate realistic anatomical structures through outpainting, effectively broadening the viewable area in medical imaging. This advancement has the potential to enhance both automatic and manual ultrasound navigation, offering a more comprehensive view that could significantly reduce the learning curve associated with ultrasound imaging and aid in more accurate diagnoses. The results confirm that echoGAN reliably reproduce detailed cardiac features, thereby promising a significant step forward in the field of non-invasive cardiac naviagation and diagnostics.
Semi-supervised ViT knowledge distillation network with style transfer normalization for colorectal liver metastases survival prediction
Nov 17, 2023



Abstract:Colorectal liver metastases (CLM) significantly impact colon cancer patients, influencing survival based on systemic chemotherapy response. Traditional methods like tumor grading scores (e.g., tumor regression grade - TRG) for prognosis suffer from subjectivity, time constraints, and expertise demands. Current machine learning approaches often focus on radiological data, yet the relevance of histological images for survival predictions, capturing intricate tumor microenvironment characteristics, is gaining recognition. To address these limitations, we propose an end-to-end approach for automated prognosis prediction using histology slides stained with H&E and HPS. We first employ a Generative Adversarial Network (GAN) for slide normalization to reduce staining variations and improve the overall quality of the images that are used as input to our prediction pipeline. We propose a semi-supervised model to perform tissue classification from sparse annotations, producing feature maps. We use an attention-based approach that weighs the importance of different slide regions in producing the final classification results. We exploit the extracted features for the metastatic nodules and surrounding tissue to train a prognosis model. In parallel, we train a vision Transformer (ViT) in a knowledge distillation framework to replicate and enhance the performance of the prognosis prediction. In our evaluation on a clinical dataset of 258 patients, our approach demonstrates superior performance with c-indexes of 0.804 (0.014) for OS and 0.733 (0.014) for TTR. Achieving 86.9% to 90.3% accuracy in predicting TRG dichotomization and 78.5% to 82.1% accuracy for the 3-class TRG classification task, our approach outperforms comparative methods. Our proposed pipeline can provide automated prognosis for pathologists and oncologists, and can greatly promote precision medicine progress in managing CLM patients.
Image-level supervision and self-training for transformer-based cross-modality tumor segmentation
Sep 17, 2023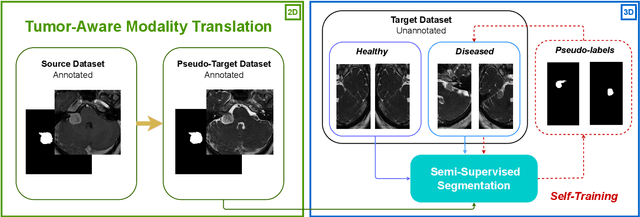
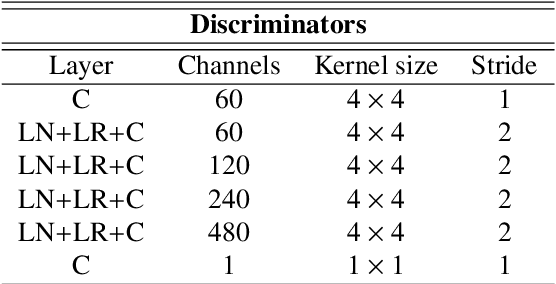
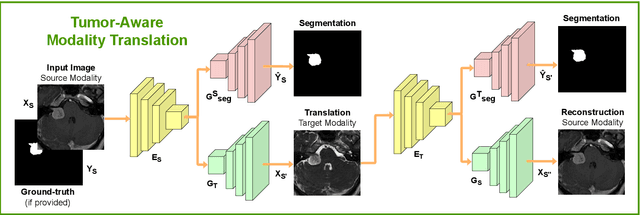
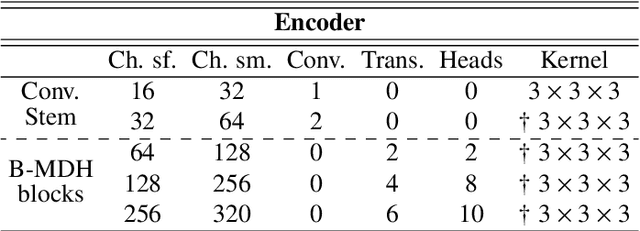
Abstract:Deep neural networks are commonly used for automated medical image segmentation, but models will frequently struggle to generalize well across different imaging modalities. This issue is particularly problematic due to the limited availability of annotated data, making it difficult to deploy these models on a larger scale. To overcome these challenges, we propose a new semi-supervised training strategy called MoDATTS. Our approach is designed for accurate cross-modality 3D tumor segmentation on unpaired bi-modal datasets. An image-to-image translation strategy between imaging modalities is used to produce annotated pseudo-target volumes and improve generalization to the unannotated target modality. We also use powerful vision transformer architectures and introduce an iterative self-training procedure to further close the domain gap between modalities. MoDATTS additionally allows the possibility to extend the training to unannotated target data by exploiting image-level labels with an unsupervised objective that encourages the model to perform 3D diseased-to-healthy translation by disentangling tumors from the background. The proposed model achieves superior performance compared to other methods from participating teams in the CrossMoDA 2022 challenge, as evidenced by its reported top Dice score of 0.87+/-0.04 for the VS segmentation. MoDATTS also yields consistent improvements in Dice scores over baselines on a cross-modality brain tumor segmentation task composed of four different contrasts from the BraTS 2020 challenge dataset, where 95% of a target supervised model performance is reached. We report that 99% and 100% of this maximum performance can be attained if 20% and 50% of the target data is additionally annotated, which further demonstrates that MoDATTS can be leveraged to reduce the annotation burden.
End-to-end Deformable Attention Graph Neural Network for Single-view Liver Mesh Reconstruction
Mar 13, 2023



Abstract:Intensity modulated radiotherapy (IMRT) is one of the most common modalities for treating cancer patients. One of the biggest challenges is precise treatment delivery that accounts for varying motion patterns originating from free-breathing. Currently, image-guided solutions for IMRT is limited to 2D guidance due to the complexity of 3D tracking solutions. We propose a novel end-to-end attention graph neural network model that generates in real-time a triangular shape of the liver based on a reference segmentation obtained at the preoperative phase and a 2D MRI coronal slice taken during the treatment. Graph neural networks work directly with graph data and can capture hidden patterns in non-Euclidean domains. Furthermore, contrary to existing methods, it produces the shape entirely in a mesh structure and correctly infers mesh shape and position based on a surrogate image. We define two on-the-fly approaches to make the correspondence of liver mesh vertices with 2D images obtained during treatment. Furthermore, we introduce a novel task-specific identity loss to constrain the deformation of the liver in the graph neural network to limit phenomenons such as flying vertices or mesh holes. The proposed method achieves results with an average error of 3.06 +- 0.7 mm and Chamfer distance with L2 norm of 63.14 +- 27.28.
Comparing 3D deformations between longitudinal daily CBCT acquisitions using CNN for head and neck radiotherapy toxicity prediction
Mar 07, 2023Abstract:Adaptive radiotherapy is a growing field of study in cancer treatment due to it's objective in sparing healthy tissue. The standard of care in several institutions includes longitudinal cone-beam computed tomography (CBCT) acquisitions to monitor changes, but have yet to be used to improve tumor control while managing side-effects. The aim of this study is to demonstrate the clinical value of pre-treatment CBCT acquired daily during radiation therapy treatment for head and neck cancers for the downstream task of predicting severe toxicity occurrence: reactive feeding tube (NG), hospitalization and radionecrosis. For this, we propose a deformable 3D classification pipeline that includes a component analyzing the Jacobian matrix of the deformation between planning CT and longitudinal CBCT, as well as clinical data. The model is based on a multi-branch 3D residual convolutional neural network, while the CT to CBCT registration is based on a pair of VoxelMorph architectures. Accuracies of 85.8% and 75.3% was found for radionecrosis and hospitalization, respectively, with similar performance as early as after the first week of treatment. For NG tube risk, performance improves with increasing the timing of the CBCT fraction, reaching 83.1% after the $5_{th}$ week of treatment.
Prediction of a T-cell/MHC-I-based immune profile for colorectal liver metastases from CT images using ensemble learning
Mar 06, 2023



Abstract:Colorectal cancer liver metastases (CLM) are the most common type of distant metastases originating from the abdomen and are characterized by a high recurrence rate after curative resection. It has been previously reported that CLM presenting a low cluster of differentiation 3 (CD3) positive T-cell infiltration density concurrent with a high major histocompatibility complex class I (MHC-I) expression were associated with poor clinical outcomes. In this study, we attempt to noninvasively predict whether a CLM exhibit the CD3LowMHCHigh immunological profile using preoperative CT images. To this end, we propose an ensemble network combining multiple Attentive Interpretable Tabular learning (TabNet) models, trained using CT-derived radiomic features. A total of 160 CLM were included in this study and randomly divided between a training set (n=130) and a hold-out test set (n=30). The proposed model yielded good prediction performance on the test set with an accuracy of 70.0% [95% confidence interval 53.6%-86.4%] and an area under the curve of 69.4% [52.9%-85.9%]. It also outperformed other off-the-shelf machine learning models. We finally demonstrated that the predicted immune profile was associated with a shorter disease-specific survival (p = .023) and time-to-recurrence (p = .020), showing the value of assessing the immune response.
M-GenSeg: Domain Adaptation For Target Modality Tumor Segmentation With Annotation-Efficient Supervision
Dec 14, 2022



Abstract:Automated medical image segmentation using deep neural networks typically requires substantial supervised training. However, these models fail to generalize well across different imaging modalities. This shortcoming, amplified by the limited availability of annotated data, has been hampering the deployment of such methods at a larger scale across modalities. To address these issues, we propose M-GenSeg, a new semi-supervised training strategy for accurate cross-modality tumor segmentation on unpaired bi-modal datasets. Based on image-level labels, a first unsupervised objective encourages the model to perform diseased to healthy translation by disentangling tumors from the background, which encompasses the segmentation task. Then, teaching the model to translate between image modalities enables the synthesis of target images from a source modality, thus leveraging the pixel-level annotations from the source modality to enforce generalization to the target modality images. We evaluated the performance on a brain tumor segmentation datasets composed of four different contrast sequences from the public BraTS 2020 challenge dataset. We report consistent improvement in Dice scores on both source and unannotated target modalities. On all twelve distinct domain adaptation experiments, the proposed model shows a clear improvement over state-of-the-art domain-adaptive baselines, with absolute Dice gains on the target modality reaching 0.15.
Label noise in segmentation networks : mitigation must deal with bias
Jul 05, 2021


Abstract:Imperfect labels limit the quality of predictions learned by deep neural networks. This is particularly relevant in medical image segmentation, where reference annotations are difficult to collect and vary significantly even across expert annotators. Prior work on mitigating label noise focused on simple models of mostly uniform noise. In this work, we explore biased and unbiased errors artificially introduced to brain tumour annotations on MRI data. We found that supervised and semi-supervised segmentation methods are robust or fairly robust to unbiased errors but sensitive to biased errors. It is therefore important to identify the sorts of errors expected in medical image labels and especially mitigate the biased errors.
3D B-mode ultrasound speckle reduction using deep learning for 3D registration applications
Aug 03, 2020



Abstract:Ultrasound (US) speckles are granular patterns which can impede image post-processing tasks, such as image segmentation and registration. Conventional filtering approaches are commonly used to remove US speckles, while their main drawback is long run-time in a 3D scenario. Although a few studies were conducted to remove 2D US speckles using deep learning, to our knowledge, there is no study to perform speckle reduction of 3D B-mode US using deep learning. In this study, we propose a 3D dense U-Net model to process 3D US B-mode data from a clinical US system. The model's results were applied to 3D registration. We show that our deep learning framework can obtain similar suppression and mean preservation index (1.066) on speckle reduction when compared to conventional filtering approaches (0.978), while reducing the runtime by two orders of magnitude. Moreover, it is found that the speckle reduction using our deep learning model contributes to improving the 3D registration performance. The mean square error of 3D registration on 3D data using 3D U-Net speckle reduction is reduced by half compared to that with speckles.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge