Image-level supervision and self-training for transformer-based cross-modality tumor segmentation
Paper and Code
Sep 17, 2023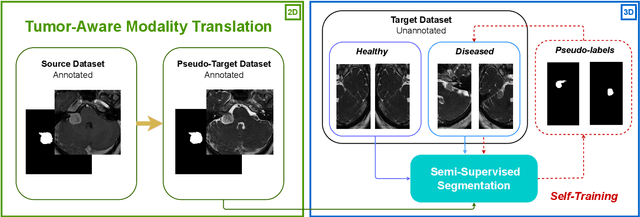
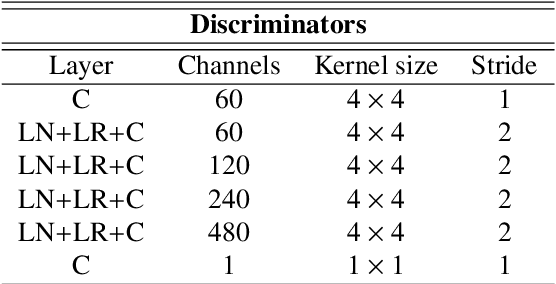
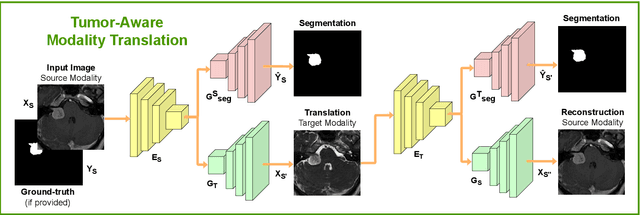
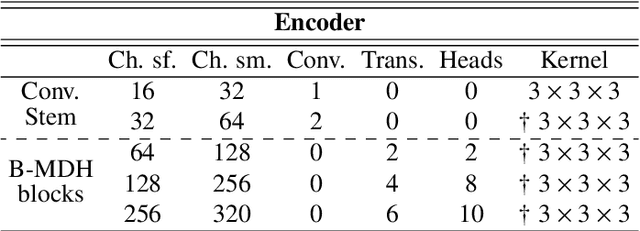
Deep neural networks are commonly used for automated medical image segmentation, but models will frequently struggle to generalize well across different imaging modalities. This issue is particularly problematic due to the limited availability of annotated data, making it difficult to deploy these models on a larger scale. To overcome these challenges, we propose a new semi-supervised training strategy called MoDATTS. Our approach is designed for accurate cross-modality 3D tumor segmentation on unpaired bi-modal datasets. An image-to-image translation strategy between imaging modalities is used to produce annotated pseudo-target volumes and improve generalization to the unannotated target modality. We also use powerful vision transformer architectures and introduce an iterative self-training procedure to further close the domain gap between modalities. MoDATTS additionally allows the possibility to extend the training to unannotated target data by exploiting image-level labels with an unsupervised objective that encourages the model to perform 3D diseased-to-healthy translation by disentangling tumors from the background. The proposed model achieves superior performance compared to other methods from participating teams in the CrossMoDA 2022 challenge, as evidenced by its reported top Dice score of 0.87+/-0.04 for the VS segmentation. MoDATTS also yields consistent improvements in Dice scores over baselines on a cross-modality brain tumor segmentation task composed of four different contrasts from the BraTS 2020 challenge dataset, where 95% of a target supervised model performance is reached. We report that 99% and 100% of this maximum performance can be attained if 20% and 50% of the target data is additionally annotated, which further demonstrates that MoDATTS can be leveraged to reduce the annotation burden.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge