William Trung Le
Semi-supervised ViT knowledge distillation network with style transfer normalization for colorectal liver metastases survival prediction
Nov 17, 2023



Abstract:Colorectal liver metastases (CLM) significantly impact colon cancer patients, influencing survival based on systemic chemotherapy response. Traditional methods like tumor grading scores (e.g., tumor regression grade - TRG) for prognosis suffer from subjectivity, time constraints, and expertise demands. Current machine learning approaches often focus on radiological data, yet the relevance of histological images for survival predictions, capturing intricate tumor microenvironment characteristics, is gaining recognition. To address these limitations, we propose an end-to-end approach for automated prognosis prediction using histology slides stained with H&E and HPS. We first employ a Generative Adversarial Network (GAN) for slide normalization to reduce staining variations and improve the overall quality of the images that are used as input to our prediction pipeline. We propose a semi-supervised model to perform tissue classification from sparse annotations, producing feature maps. We use an attention-based approach that weighs the importance of different slide regions in producing the final classification results. We exploit the extracted features for the metastatic nodules and surrounding tissue to train a prognosis model. In parallel, we train a vision Transformer (ViT) in a knowledge distillation framework to replicate and enhance the performance of the prognosis prediction. In our evaluation on a clinical dataset of 258 patients, our approach demonstrates superior performance with c-indexes of 0.804 (0.014) for OS and 0.733 (0.014) for TTR. Achieving 86.9% to 90.3% accuracy in predicting TRG dichotomization and 78.5% to 82.1% accuracy for the 3-class TRG classification task, our approach outperforms comparative methods. Our proposed pipeline can provide automated prognosis for pathologists and oncologists, and can greatly promote precision medicine progress in managing CLM patients.
Image-level supervision and self-training for transformer-based cross-modality tumor segmentation
Sep 17, 2023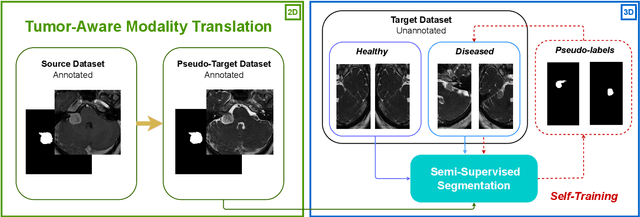
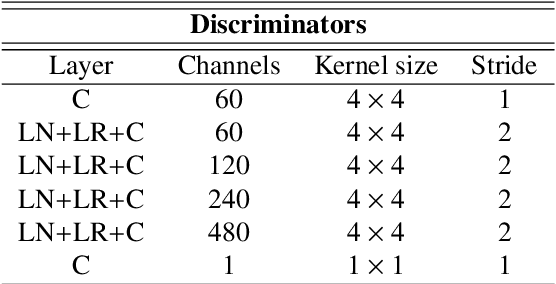
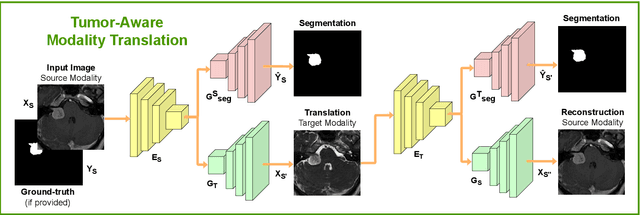
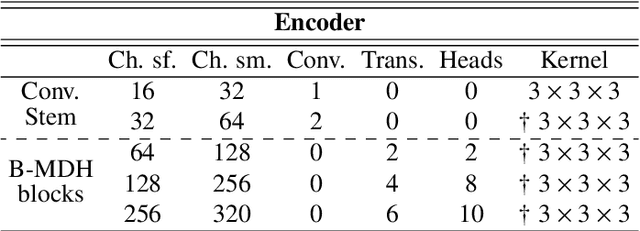
Abstract:Deep neural networks are commonly used for automated medical image segmentation, but models will frequently struggle to generalize well across different imaging modalities. This issue is particularly problematic due to the limited availability of annotated data, making it difficult to deploy these models on a larger scale. To overcome these challenges, we propose a new semi-supervised training strategy called MoDATTS. Our approach is designed for accurate cross-modality 3D tumor segmentation on unpaired bi-modal datasets. An image-to-image translation strategy between imaging modalities is used to produce annotated pseudo-target volumes and improve generalization to the unannotated target modality. We also use powerful vision transformer architectures and introduce an iterative self-training procedure to further close the domain gap between modalities. MoDATTS additionally allows the possibility to extend the training to unannotated target data by exploiting image-level labels with an unsupervised objective that encourages the model to perform 3D diseased-to-healthy translation by disentangling tumors from the background. The proposed model achieves superior performance compared to other methods from participating teams in the CrossMoDA 2022 challenge, as evidenced by its reported top Dice score of 0.87+/-0.04 for the VS segmentation. MoDATTS also yields consistent improvements in Dice scores over baselines on a cross-modality brain tumor segmentation task composed of four different contrasts from the BraTS 2020 challenge dataset, where 95% of a target supervised model performance is reached. We report that 99% and 100% of this maximum performance can be attained if 20% and 50% of the target data is additionally annotated, which further demonstrates that MoDATTS can be leveraged to reduce the annotation burden.
Comparing 3D deformations between longitudinal daily CBCT acquisitions using CNN for head and neck radiotherapy toxicity prediction
Mar 07, 2023Abstract:Adaptive radiotherapy is a growing field of study in cancer treatment due to it's objective in sparing healthy tissue. The standard of care in several institutions includes longitudinal cone-beam computed tomography (CBCT) acquisitions to monitor changes, but have yet to be used to improve tumor control while managing side-effects. The aim of this study is to demonstrate the clinical value of pre-treatment CBCT acquired daily during radiation therapy treatment for head and neck cancers for the downstream task of predicting severe toxicity occurrence: reactive feeding tube (NG), hospitalization and radionecrosis. For this, we propose a deformable 3D classification pipeline that includes a component analyzing the Jacobian matrix of the deformation between planning CT and longitudinal CBCT, as well as clinical data. The model is based on a multi-branch 3D residual convolutional neural network, while the CT to CBCT registration is based on a pair of VoxelMorph architectures. Accuracies of 85.8% and 75.3% was found for radionecrosis and hospitalization, respectively, with similar performance as early as after the first week of treatment. For NG tube risk, performance improves with increasing the timing of the CBCT fraction, reaching 83.1% after the $5_{th}$ week of treatment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge