Francisco Perdigon Romero
Attention-based Class-Conditioned Alignment for Multi-Source Domain Adaptive Object Detection
Mar 14, 2024



Abstract:Domain adaptation methods for object detection (OD) strive to mitigate the impact of distribution shifts by promoting feature alignment across source and target domains. Multi-source domain adaptation (MSDA) allows leveraging multiple annotated source datasets, and unlabeled target data to improve the accuracy and robustness of the detection model. Most state-of-the-art MSDA methods for OD perform feature alignment in a class-agnostic manner. This is challenging since the objects have unique modal information due to variations in object appearance across domains. A recent prototype-based approach proposed a class-wise alignment, yet it suffers from error accumulation due to noisy pseudo-labels which can negatively affect adaptation with imbalanced data. To overcome these limitations, we propose an attention-based class-conditioned alignment scheme for MSDA that aligns instances of each object category across domains. In particular, an attention module coupled with an adversarial domain classifier allows learning domain-invariant and class-specific instance representations. Experimental results on multiple benchmarking MSDA datasets indicate that our method outperforms the state-of-the-art methods and is robust to class imbalance. Our code is available at https://github.com/imatif17/ACIA.
Semi-supervised ViT knowledge distillation network with style transfer normalization for colorectal liver metastases survival prediction
Nov 17, 2023



Abstract:Colorectal liver metastases (CLM) significantly impact colon cancer patients, influencing survival based on systemic chemotherapy response. Traditional methods like tumor grading scores (e.g., tumor regression grade - TRG) for prognosis suffer from subjectivity, time constraints, and expertise demands. Current machine learning approaches often focus on radiological data, yet the relevance of histological images for survival predictions, capturing intricate tumor microenvironment characteristics, is gaining recognition. To address these limitations, we propose an end-to-end approach for automated prognosis prediction using histology slides stained with H&E and HPS. We first employ a Generative Adversarial Network (GAN) for slide normalization to reduce staining variations and improve the overall quality of the images that are used as input to our prediction pipeline. We propose a semi-supervised model to perform tissue classification from sparse annotations, producing feature maps. We use an attention-based approach that weighs the importance of different slide regions in producing the final classification results. We exploit the extracted features for the metastatic nodules and surrounding tissue to train a prognosis model. In parallel, we train a vision Transformer (ViT) in a knowledge distillation framework to replicate and enhance the performance of the prognosis prediction. In our evaluation on a clinical dataset of 258 patients, our approach demonstrates superior performance with c-indexes of 0.804 (0.014) for OS and 0.733 (0.014) for TTR. Achieving 86.9% to 90.3% accuracy in predicting TRG dichotomization and 78.5% to 82.1% accuracy for the 3-class TRG classification task, our approach outperforms comparative methods. Our proposed pipeline can provide automated prognosis for pathologists and oncologists, and can greatly promote precision medicine progress in managing CLM patients.
Multi-Source Domain Adaptation for Object Detection with Prototype-based Mean-teacher
Sep 26, 2023



Abstract:Adapting visual object detectors to operational target domains is a challenging task, commonly achieved using unsupervised domain adaptation (UDA) methods. When the labeled dataset is coming from multiple source domains, treating them as separate domains and performing a multi-source domain adaptation (MSDA) improves the accuracy and robustness over mixing these source domains and performing a UDA, as observed by recent studies in MSDA. Existing MSDA methods learn domain invariant and domain-specific parameters (for each source domain) for the adaptation. However, unlike single-source UDA methods, learning domain-specific parameters makes them grow significantly proportional to the number of source domains used. This paper proposes a novel MSDA method called Prototype-based Mean-Teacher (PMT), which uses class prototypes instead of domain-specific subnets to preserve domain-specific information. These prototypes are learned using a contrastive loss, aligning the same categories across domains and separating different categories far apart. Because of the use of prototypes, the parameter size of our method does not increase significantly with the number of source domains, thus reducing memory issues and possible overfitting. Empirical studies show PMT outperforms state-of-the-art MSDA methods on several challenging object detection datasets.
Semi-Weakly Supervised Object Detection by Sampling Pseudo Ground-Truth Boxes
Apr 01, 2022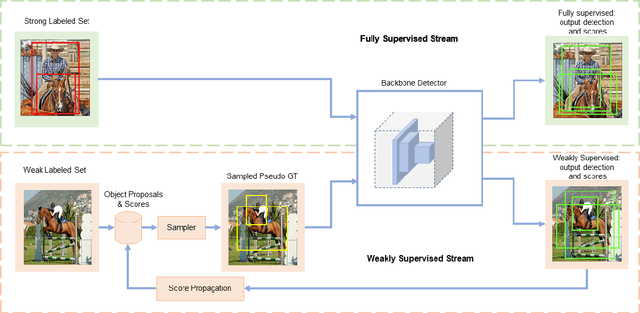
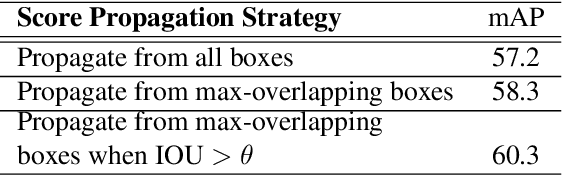
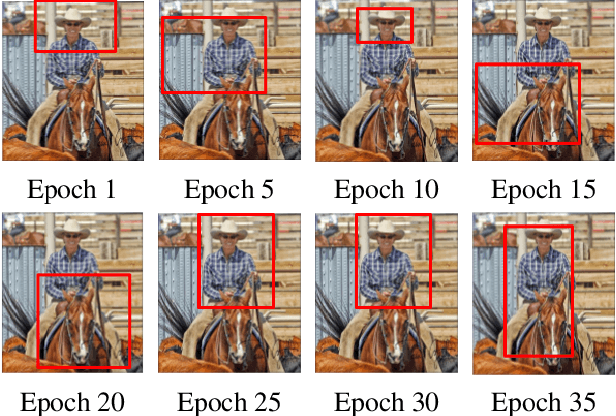

Abstract:Semi- and weakly-supervised learning have recently attracted considerable attention in the object detection literature since they can alleviate the cost of annotation needed to successfully train deep learning models. State-of-art approaches for semi-supervised learning rely on student-teacher models trained using a multi-stage process, and considerable data augmentation. Custom networks have been developed for the weakly-supervised setting, making it difficult to adapt to different detectors. In this paper, a weakly semi-supervised training method is introduced that reduces these training challenges, yet achieves state-of-the-art performance by leveraging only a small fraction of fully-labeled images with information in weakly-labeled images. In particular, our generic sampling-based learning strategy produces pseudo-ground-truth (GT) bounding box annotations in an online fashion, eliminating the need for multi-stage training, and student-teacher network configurations. These pseudo GT boxes are sampled from weakly-labeled images based on the categorical score of object proposals accumulated via a score propagation process. Empirical results on the Pascal VOC dataset, indicate that the proposed approach improves performance by 5.0% when using VOC 2007 as fully-labeled, and VOC 2012 as weak-labeled data. Also, with 5-10% fully annotated images, we observed an improvement of more than 10% in mAP, showing that a modest investment in image-level annotation, can substantially improve detection performance.
DeepFilter: an ECG baseline wander removal filter using deep learning techniques
Jan 12, 2021

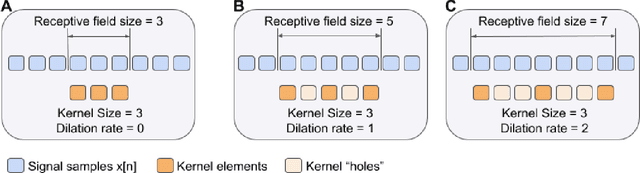
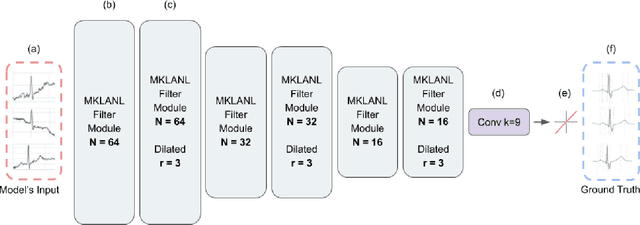
Abstract:According to the World Health Organization, around 36% of the annual deaths are associated with cardiovascular diseases and 90% of heart attacks are preventable. Electrocardiogram signal analysis in ambulatory electrocardiography, during an exercise stress test, and in resting conditions allows cardiovascular disease diagnosis. However, during the acquisition, there is a variety of noises that may damage the signal quality thereby compromising their diagnostic potential. The baseline wander is one of the most undesirable noises. In this work, we propose a novel algorithm for BLW noise filtering using deep learning techniques. The model performance was validated using the QT Database and the MIT-BIH Noise Stress Test Database from Physionet. In addition, several comparative experiments were performed against state-of-the-art methods using traditional filtering procedures as well as deep learning techniques. The proposed approach yields the best results on four similarity metrics: the sum of squared distance, maximum absolute square, percentage of root distance, and cosine similarity with 4.29 (6.35) au, 0.34 (0.25) au, 45.35 (29.69) au and, 91.46 (8.61) au, respectively. The source code of this work, containing our method and related implementations, is freely available on Github.
Spine intervertebral disc labeling using a fully convolutional redundant counting model
Mar 11, 2020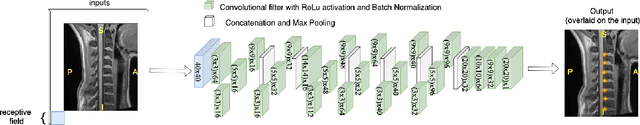
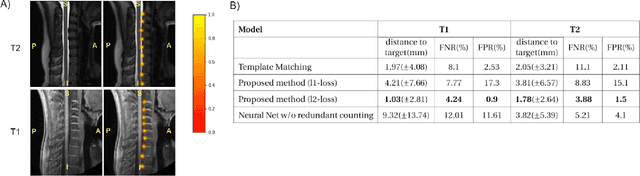
Abstract:Labeling intervertebral discs is relevant as it notably enables clinicians to understand the relationship between a patient's symptoms (pain, paralysis) and the exact level of spinal cord injury. However manually labeling those discs is a tedious and user-biased task which would benefit from automated methods. While some automated methods already exist for MRI and CT-scan, they are either not publicly available, or fail to generalize across various imaging contrasts. In this paper we combine a Fully Convolutional Network (FCN) with inception modules to localize and label intervertebral discs. We demonstrate a proof-of-concept application in a publicly-available multi-center and multi-contrast MRI database (n=235 subjects). The code is publicly available at https://github.com/neuropoly/vertebral-labeling-deep-learning.
End-to-End Discriminative Deep Network for Liver Lesion Classification
Jan 28, 2019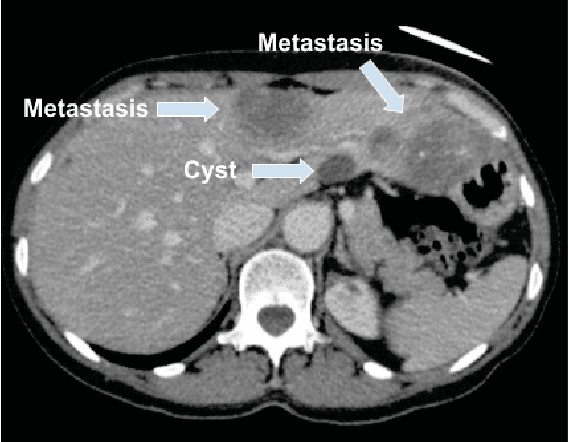
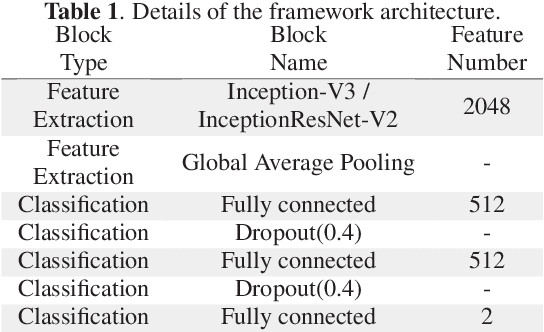
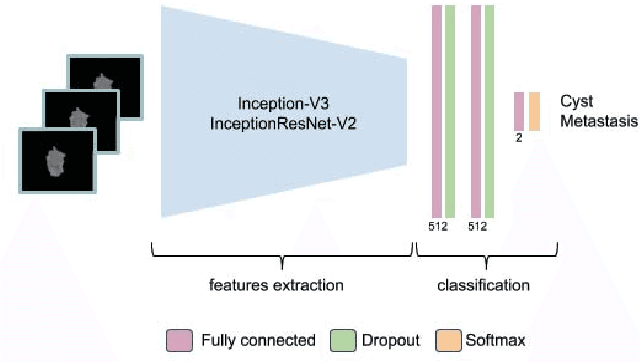
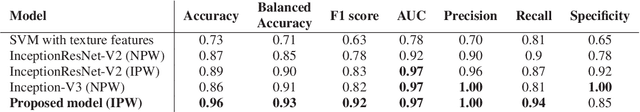
Abstract:Colorectal liver metastasis is one of most aggressive liver malignancies. While the definition of lesion type based on CT images determines the diagnosis and therapeutic strategy, the discrimination between cancerous and non-cancerous lesions are critical and requires highly skilled expertise, experience and time. In the present work we introduce an end-to-end deep learning approach to assist in the discrimination between liver metastases from colorectal cancer and benign cysts in abdominal CT images of the liver. Our approach incorporates the efficient feature extraction of InceptionV3 combined with residual connections and pre-trained weights from ImageNet. The architecture also includes fully connected classification layers to generate a probabilistic output of lesion type. We use an in-house clinical biobank with 230 liver lesions originating from 63 patients. With an accuracy of 0.96 and a F1-score of 0.92, the results obtained with the proposed approach surpasses state of the art methods. Our work provides the basis for incorporating machine learning tools in specialized radiology software to assist physicians in the early detection and treatment of liver lesions.
Multi-Level Batch Normalization In Deep Networks For Invasive Ductal Carcinoma Cell Discrimination In Histopathology Images
Jan 11, 2019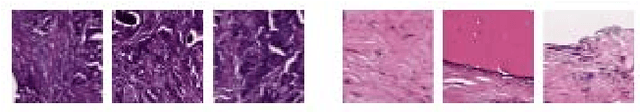
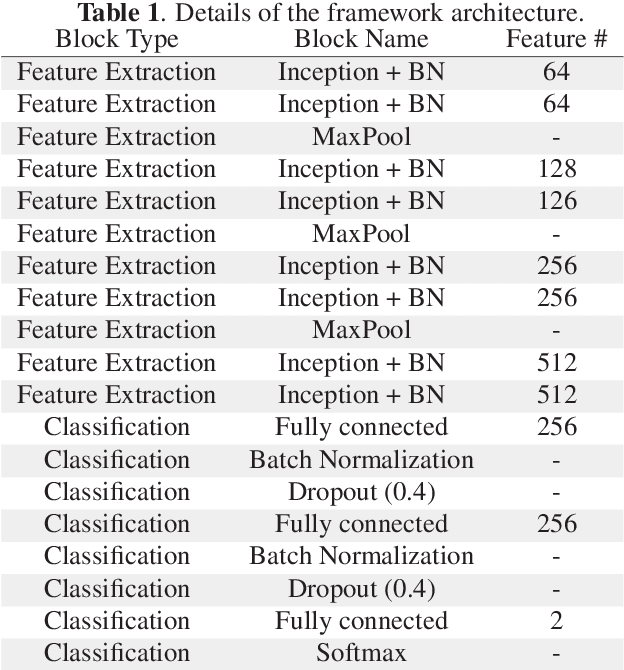
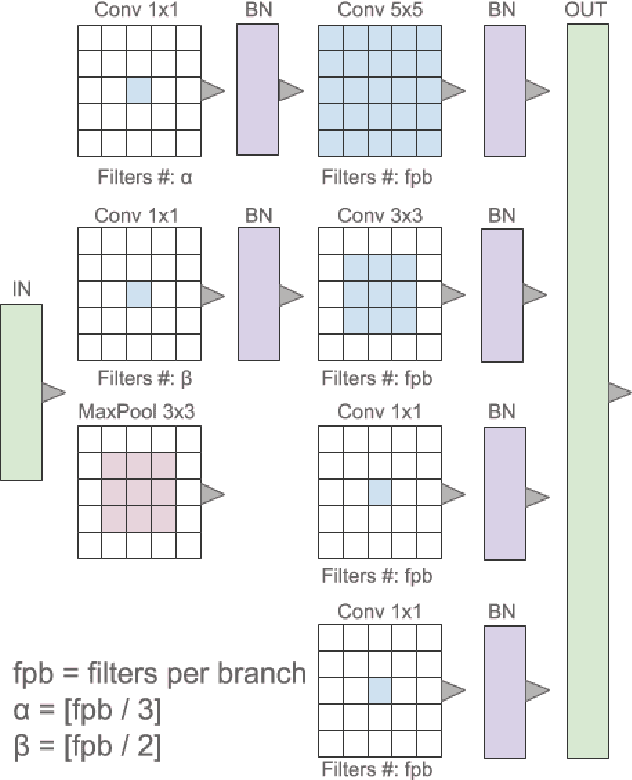
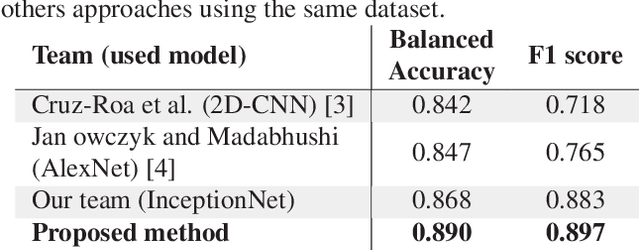
Abstract:Breast cancer is the most diagnosed cancer and the most predominant cause of death in women worldwide. Imaging techniques such as the breast cancer pathology helps in the diagnosis and monitoring of the disease. However identification of malignant cells can be challenging given the high heterogeneity in tissue absorbotion from staining agents. In this work, we present a novel approach for Invasive Ductal Carcinoma (IDC) cells discrimination in histopathology slides. We propose a model derived from the Inception architecture, proposing a multi-level batch normalization module between each convolutional steps. This module was used as a base block for the feature extraction in a CNN architecture. We used the open IDC dataset in which we obtained a balanced accuracy of 0.89 and an F1 score of 0.90, thus surpassing recent state of the art classification algorithms tested on this public dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge