Simon Turcotte
Semi-supervised ViT knowledge distillation network with style transfer normalization for colorectal liver metastases survival prediction
Nov 17, 2023



Abstract:Colorectal liver metastases (CLM) significantly impact colon cancer patients, influencing survival based on systemic chemotherapy response. Traditional methods like tumor grading scores (e.g., tumor regression grade - TRG) for prognosis suffer from subjectivity, time constraints, and expertise demands. Current machine learning approaches often focus on radiological data, yet the relevance of histological images for survival predictions, capturing intricate tumor microenvironment characteristics, is gaining recognition. To address these limitations, we propose an end-to-end approach for automated prognosis prediction using histology slides stained with H&E and HPS. We first employ a Generative Adversarial Network (GAN) for slide normalization to reduce staining variations and improve the overall quality of the images that are used as input to our prediction pipeline. We propose a semi-supervised model to perform tissue classification from sparse annotations, producing feature maps. We use an attention-based approach that weighs the importance of different slide regions in producing the final classification results. We exploit the extracted features for the metastatic nodules and surrounding tissue to train a prognosis model. In parallel, we train a vision Transformer (ViT) in a knowledge distillation framework to replicate and enhance the performance of the prognosis prediction. In our evaluation on a clinical dataset of 258 patients, our approach demonstrates superior performance with c-indexes of 0.804 (0.014) for OS and 0.733 (0.014) for TTR. Achieving 86.9% to 90.3% accuracy in predicting TRG dichotomization and 78.5% to 82.1% accuracy for the 3-class TRG classification task, our approach outperforms comparative methods. Our proposed pipeline can provide automated prognosis for pathologists and oncologists, and can greatly promote precision medicine progress in managing CLM patients.
Prediction of a T-cell/MHC-I-based immune profile for colorectal liver metastases from CT images using ensemble learning
Mar 06, 2023



Abstract:Colorectal cancer liver metastases (CLM) are the most common type of distant metastases originating from the abdomen and are characterized by a high recurrence rate after curative resection. It has been previously reported that CLM presenting a low cluster of differentiation 3 (CD3) positive T-cell infiltration density concurrent with a high major histocompatibility complex class I (MHC-I) expression were associated with poor clinical outcomes. In this study, we attempt to noninvasively predict whether a CLM exhibit the CD3LowMHCHigh immunological profile using preoperative CT images. To this end, we propose an ensemble network combining multiple Attentive Interpretable Tabular learning (TabNet) models, trained using CT-derived radiomic features. A total of 160 CLM were included in this study and randomly divided between a training set (n=130) and a hold-out test set (n=30). The proposed model yielded good prediction performance on the test set with an accuracy of 70.0% [95% confidence interval 53.6%-86.4%] and an area under the curve of 69.4% [52.9%-85.9%]. It also outperformed other off-the-shelf machine learning models. We finally demonstrated that the predicted immune profile was associated with a shorter disease-specific survival (p = .023) and time-to-recurrence (p = .020), showing the value of assessing the immune response.
End-to-End Discriminative Deep Network for Liver Lesion Classification
Jan 28, 2019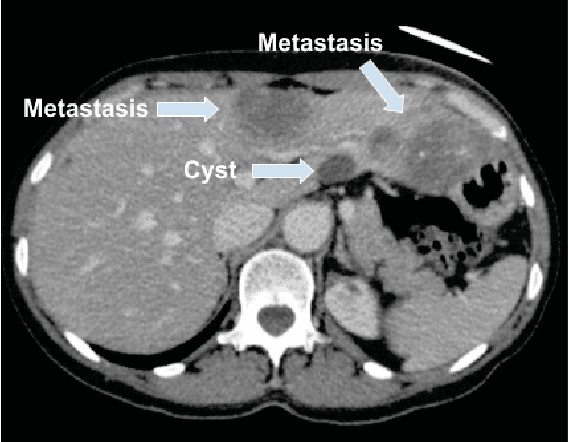
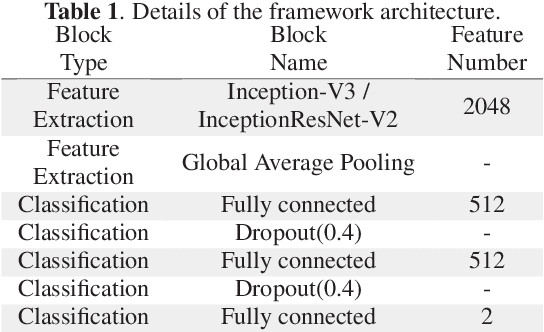
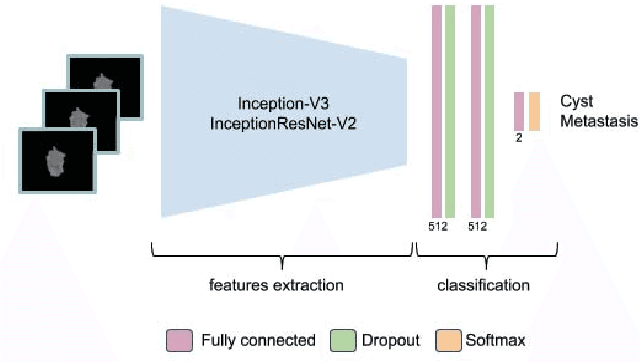
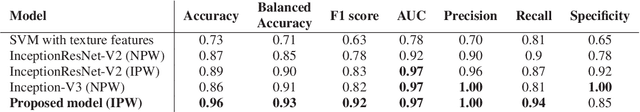
Abstract:Colorectal liver metastasis is one of most aggressive liver malignancies. While the definition of lesion type based on CT images determines the diagnosis and therapeutic strategy, the discrimination between cancerous and non-cancerous lesions are critical and requires highly skilled expertise, experience and time. In the present work we introduce an end-to-end deep learning approach to assist in the discrimination between liver metastases from colorectal cancer and benign cysts in abdominal CT images of the liver. Our approach incorporates the efficient feature extraction of InceptionV3 combined with residual connections and pre-trained weights from ImageNet. The architecture also includes fully connected classification layers to generate a probabilistic output of lesion type. We use an in-house clinical biobank with 230 liver lesions originating from 63 patients. With an accuracy of 0.96 and a F1-score of 0.92, the results obtained with the proposed approach surpasses state of the art methods. Our work provides the basis for incorporating machine learning tools in specialized radiology software to assist physicians in the early detection and treatment of liver lesions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge