Matej Gazda
Large-scale modality-invariant foundation models for brain MRI analysis: Application to lesion segmentation
Nov 14, 2025Abstract:The field of computer vision is undergoing a paradigm shift toward large-scale foundation model pre-training via self-supervised learning (SSL). Leveraging large volumes of unlabeled brain MRI data, such models can learn anatomical priors that improve few-shot performance in diverse neuroimaging tasks. However, most SSL frameworks are tailored to natural images, and their adaptation to capture multi-modal MRI information remains underexplored. This work proposes a modality-invariant representation learning setup and evaluates its effectiveness in stroke and epilepsy lesion segmentation, following large-scale pre-training. Experimental results suggest that despite successful cross-modality alignment, lesion segmentation primarily benefits from preserving fine-grained modality-specific features. Model checkpoints and code are made publicly available.
Generative Adversarial Networks in Ultrasound Imaging: Extending Field of View Beyond Conventional Limits
May 31, 2024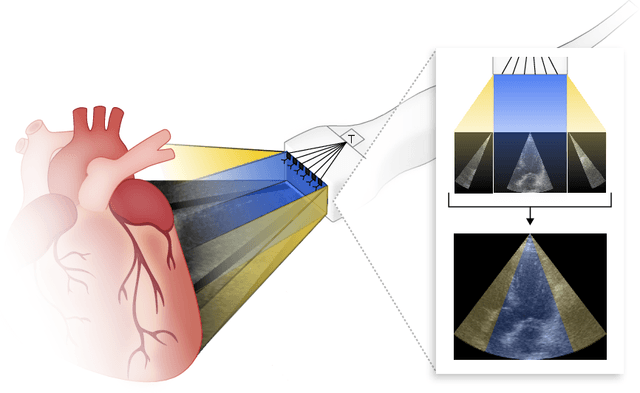
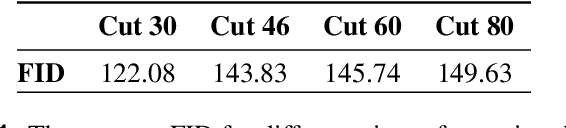
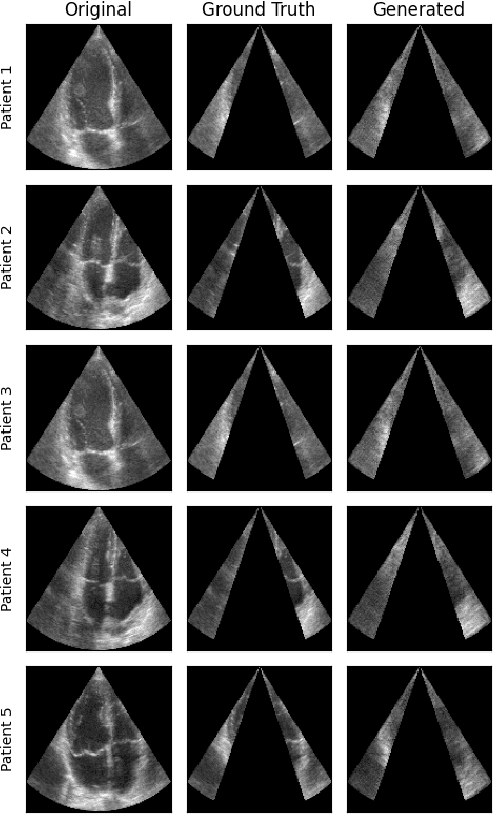
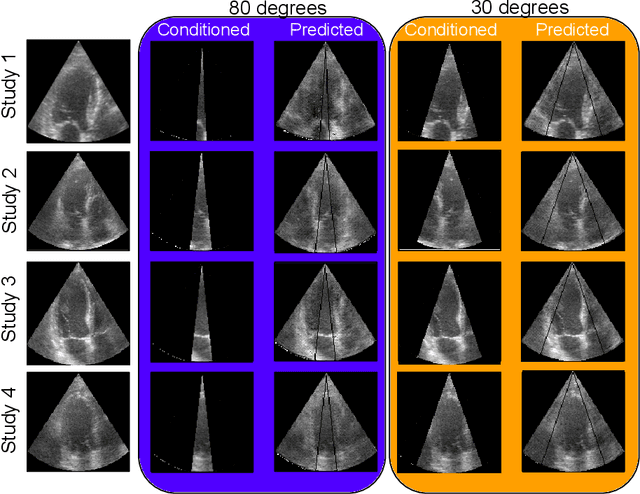
Abstract:Transthoracic Echocardiography (TTE) is a fundamental, non-invasive diagnostic tool in cardiovascular medicine, enabling detailed visualization of cardiac structures crucial for diagnosing various heart conditions. Despite its widespread use, TTE ultrasound imaging faces inherent limitations, notably the trade-off between field of view (FoV) and resolution. This paper introduces a novel application of conditional Generative Adversarial Networks (cGANs), specifically designed to extend the FoV in TTE ultrasound imaging while maintaining high resolution. Our proposed cGAN architecture, termed echoGAN, demonstrates the capability to generate realistic anatomical structures through outpainting, effectively broadening the viewable area in medical imaging. This advancement has the potential to enhance both automatic and manual ultrasound navigation, offering a more comprehensive view that could significantly reduce the learning curve associated with ultrasound imaging and aid in more accurate diagnoses. The results confirm that echoGAN reliably reproduce detailed cardiac features, thereby promising a significant step forward in the field of non-invasive cardiac naviagation and diagnostics.
End-to-end Deformable Attention Graph Neural Network for Single-view Liver Mesh Reconstruction
Mar 13, 2023



Abstract:Intensity modulated radiotherapy (IMRT) is one of the most common modalities for treating cancer patients. One of the biggest challenges is precise treatment delivery that accounts for varying motion patterns originating from free-breathing. Currently, image-guided solutions for IMRT is limited to 2D guidance due to the complexity of 3D tracking solutions. We propose a novel end-to-end attention graph neural network model that generates in real-time a triangular shape of the liver based on a reference segmentation obtained at the preoperative phase and a 2D MRI coronal slice taken during the treatment. Graph neural networks work directly with graph data and can capture hidden patterns in non-Euclidean domains. Furthermore, contrary to existing methods, it produces the shape entirely in a mesh structure and correctly infers mesh shape and position based on a surrogate image. We define two on-the-fly approaches to make the correspondence of liver mesh vertices with 2D images obtained during treatment. Furthermore, we introduce a novel task-specific identity loss to constrain the deformation of the liver in the graph neural network to limit phenomenons such as flying vertices or mesh holes. The proposed method achieves results with an average error of 3.06 +- 0.7 mm and Chamfer distance with L2 norm of 63.14 +- 27.28.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Self-supervised deep convolutional neural network for chest X-ray classification
Mar 05, 2021

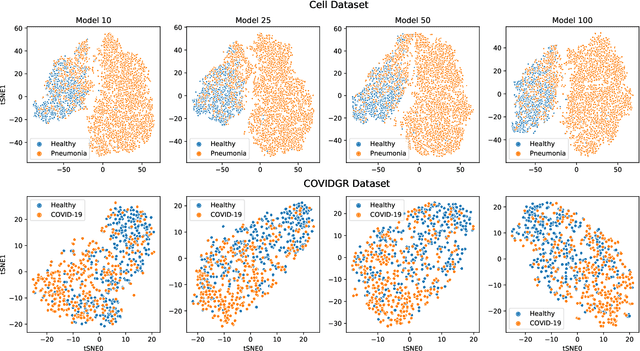
Abstract:Chest radiography is a relatively cheap, widely available medical procedure that conveys key information for making diagnostic decisions. Chest X-rays are almost always used in the diagnosis of respiratory diseases such as pneumonia or the recent COVID-19. In this paper, we propose a self-supervised deep neural network that is pretrained on an unlabeled chest X-ray dataset. The learned representations are transferred to downstream task - the classification of respiratory diseases. The results obtained on four public datasets show that our approach yields competitive results without requiring large amounts of labeled training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge