Robin Strand
Leveraging point annotations in segmentation learning with boundary loss
Nov 06, 2023Abstract:This paper investigates the combination of intensity-based distance maps with boundary loss for point-supervised semantic segmentation. By design the boundary loss imposes a stronger penalty on the false positives the farther away from the object they occur. Hence it is intuitively inappropriate for weak supervision, where the ground truth label may be much smaller than the actual object and a certain amount of false positives (w.r.t. the weak ground truth) is actually desirable. Using intensity-aware distances instead may alleviate this drawback, allowing for a certain amount of false positives without a significant increase to the training loss. The motivation for applying the boundary loss directly under weak supervision lies in its great success for fully supervised segmentation tasks, but also in not requiring extra priors or outside information that is usually required -- in some form -- with existing weakly supervised methods in the literature. This formulation also remains potentially more attractive than existing CRF-based regularizers, due to its simplicity and computational efficiency. We perform experiments on two multi-class datasets; ACDC (heart segmentation) and POEM (whole-body abdominal organ segmentation). Preliminary results are encouraging and show that this supervision strategy has great potential. On ACDC it outperforms the CRF-loss based approach, and on POEM data it performs on par with it. The code for all our experiments is openly available.
MIMIR: Deep Regression for Automated Analysis of UK Biobank Body MRI
Jun 22, 2021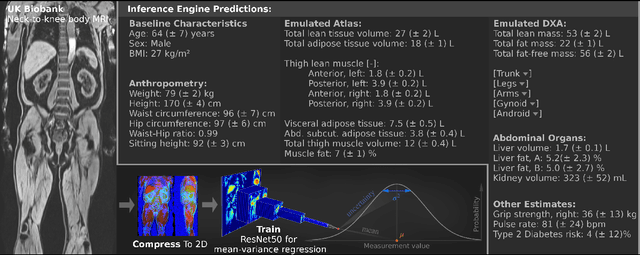
Abstract:UK Biobank (UKB) is conducting a large-scale study of more than half a million volunteers, collecting health-related information on genetics, lifestyle, blood biochemistry, and more. Medical imaging furthermore targets 100,000 subjects, with 70,000 follow-up sessions, enabling measurements of organs, muscle, and body composition. With up to 170,000 mounting MR images, various methodologies are accordingly engaged in large-scale image analysis. This work presents an experimental inference engine that can automatically predict a comprehensive profile of subject metadata from UKB neck-to-knee body MRI. In cross-validation, it accurately inferred baseline characteristics such as age, height, weight, and sex, but also emulated measurements of body composition by DXA, organ volumes, and abstract properties like grip strength, pulse rate, and type 2 diabetic status (AUC: 0.866). The proposed system can automatically analyze thousands of subjects within hours and provide individual confidence intervals. The underlying methodology is based on convolutional neural networks for image-based mean-variance regression on two-dimensional representations of the MRI data. This work aims to make the proposed system available for free to researchers, who can use it to obtain fast and fully-automated estimates of 72 different measurements immediately upon release of new UK Biobank image data.
Deep regression for uncertainty-aware and interpretable analysis of large-scale body MRI
May 17, 2021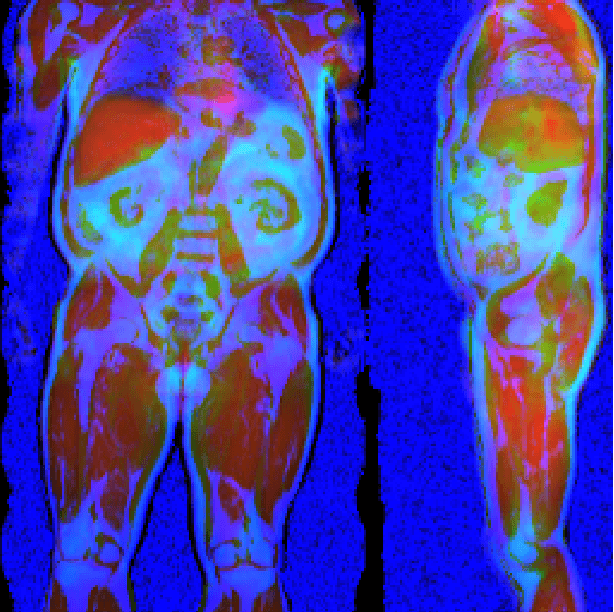
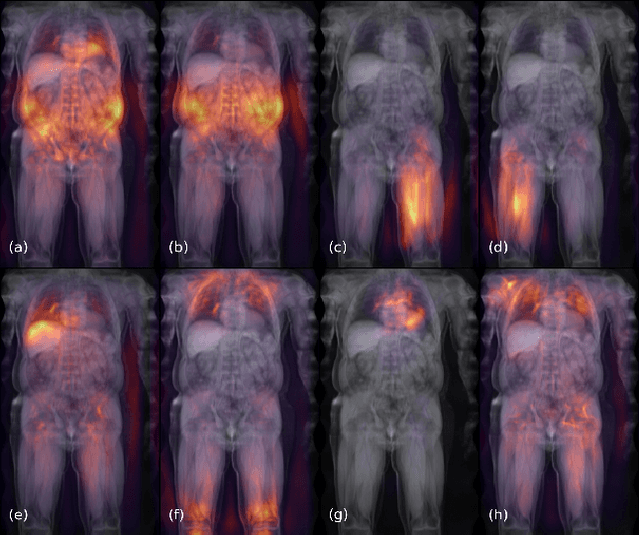
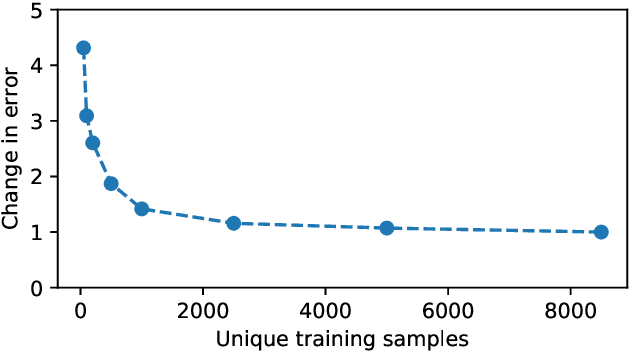
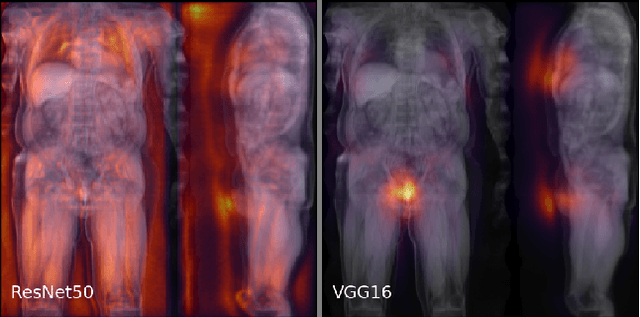
Abstract:Large-scale medical studies such as the UK Biobank examine thousands of volunteer participants with medical imaging techniques. Combined with the vast amount of collected metadata, anatomical information from these images has the potential for medical analyses at unprecedented scale. However, their evaluation often requires manual input and long processing times, limiting the amount of reference values for biomarkers and other measurements available for research. Recent approaches with convolutional neural networks for regression can perform these evaluations automatically. On magnetic resonance imaging (MRI) data of more than 40,000 UK Biobank subjects, these systems can estimate human age, body composition and more. This style of analysis is almost entirely data-driven and no manual intervention or guidance with manually segmented ground truth images is required. The networks often closely emulate the reference method that provided their training data and can reach levels of agreement comparable to the expected variability between established medical gold standard techniques. The risk of silent failure can be individually quantified by predictive uncertainty obtained from a mean-variance criterion and ensembling. Saliency analysis furthermore enables an interpretation of the underlying relevant image features and showed that the networks learned to correctly target specific organs, limbs, and regions of interest.
Uncertainty-Aware Body Composition Analysis with Deep Regression Ensembles on UK Biobank MRI
Jan 18, 2021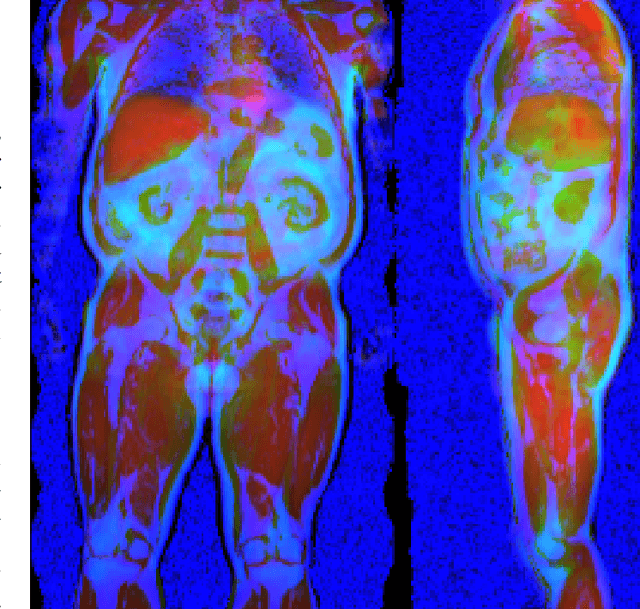
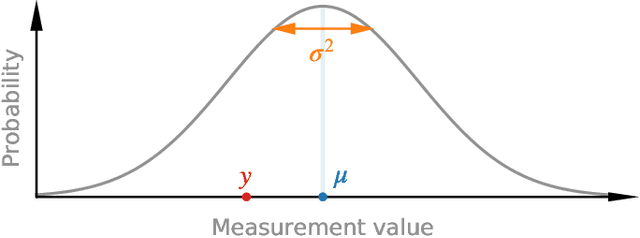
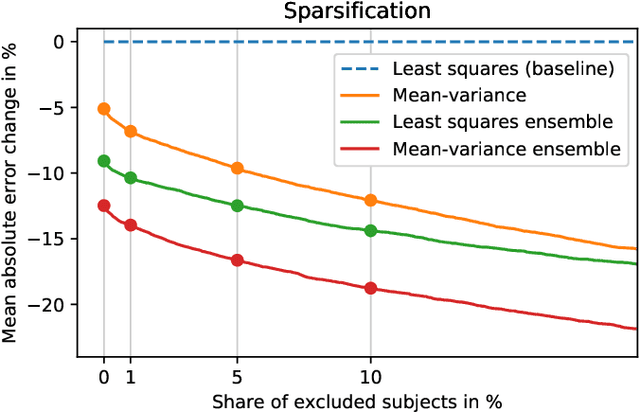
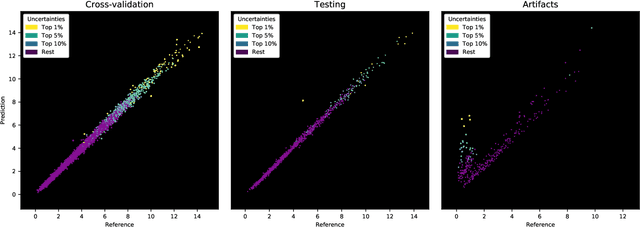
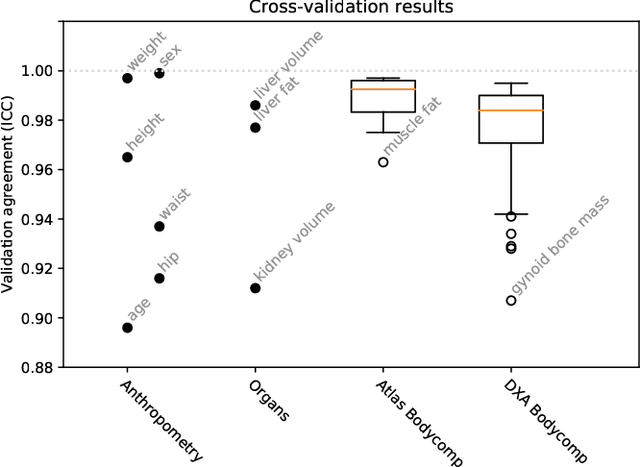
Abstract:Purpose: To enable fast and automated analysis of body composition from UK Biobank MRI with accurate estimates of individual measurement errors. Methods: In an ongoing large-scale imaging study the UK Biobank has acquired MRI of over 40,000 men and women aged 44-82. Phenotypes derived from these images, such as body composition, can reveal new links between genetics, cardiovascular disease, and metabolic conditions. In this retrospective study, neural networks were trained to provide six measurements of body composition from UK Biobank neck-to-knee body MRI. A ResNet50 architecture can automatically predict these values by image-based regression, but may also produce erroneous outliers. Predictive uncertainty, which could identify these failure cases, was therefore modeled with a mean-variance loss and ensembling. Its estimates of individual prediction errors were evaluated in cross-validation on over 8,000 subjects, tested on another 1,000 cases, and finally applied for inference. Results: Relative measurement errors below 5\% were achieved on all but one target, for intra-class correlation coefficients (ICC) above 0.97 both in validation and testing. Both mean-variance loss and ensembling yielded improvements and provided uncertainty estimates that highlighted some of the worst outlier predictions. Combined, they reached the highest quality, but also exhibited a consistent bias towards high uncertainty in heavyweight subjects. Conclusion: Mean-variance regression and ensembling provided complementary benefits for automated body composition measurements from UK Biobank MRI, reaching high speed and accuracy. These values were inferred for the entire cohort, with uncertainty estimates that can approximate the measurement errors and identify some of the worst outliers automatically.
Large-scale inference of liver fat with neural networks on UK Biobank body MRI
Jun 30, 2020
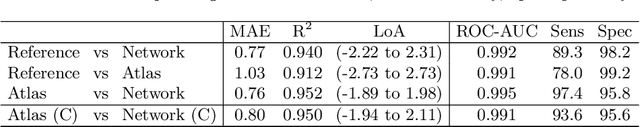
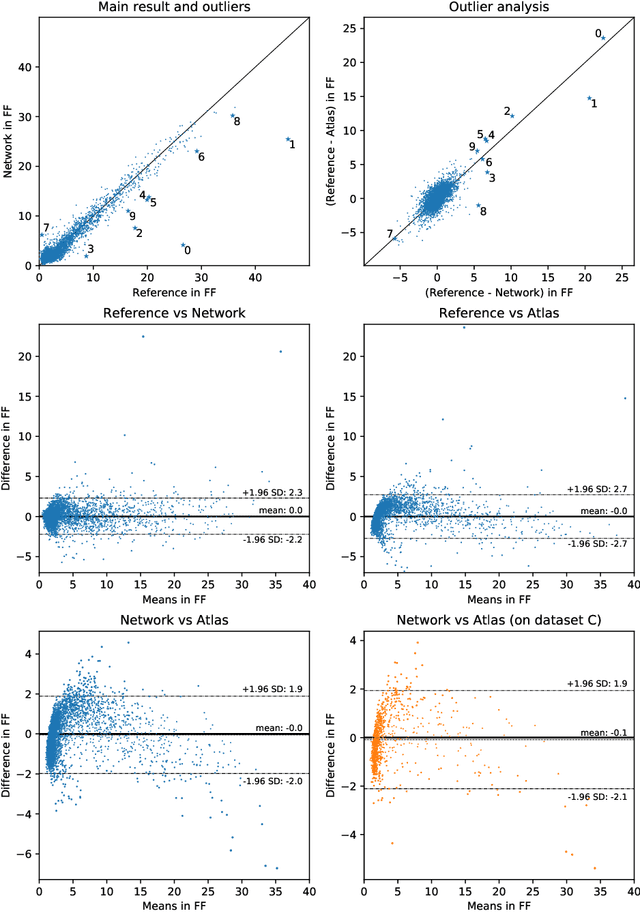
Abstract:The UK Biobank Imaging Study has acquired medical scans of more than 40,000 volunteer participants. The resulting wealth of anatomical information has been made available for research, together with extensive metadata including measurements of liver fat. These values play an important role in metabolic disease, but are only available for a minority of imaged subjects as their collection requires the careful work of image analysts on dedicated liver MRI. Another UK Biobank protocol is neck-to-knee body MRI for analysis of body composition. The resulting volumes can also quantify fat fractions, even though they were reconstructed with a two- instead of a three-point Dixon technique. In this work, a novel framework for automated inference of liver fat from UK Biobank neck-to-knee body MRI is proposed. A ResNet50 was trained for regression on two-dimensional slices from these scans and the reference values as target, without any need for ground truth segmentations. Once trained, it performs fast, objective, and fully automated predictions that require no manual intervention. On the given data, it closely emulates the reference method, reaching a level of agreement comparable to different gold standard techniques. The network learned to rectify non-linearities in the fat fraction values and identified several outliers in the reference. It outperformed a multi-atlas segmentation baseline and inferred new estimates for all imaged subjects lacking reference values, expanding the total number of liver fat measurements by factor six.
Kidney segmentation in neck-to-knee body MRI of 40,000 UK Biobank participants
Jun 12, 2020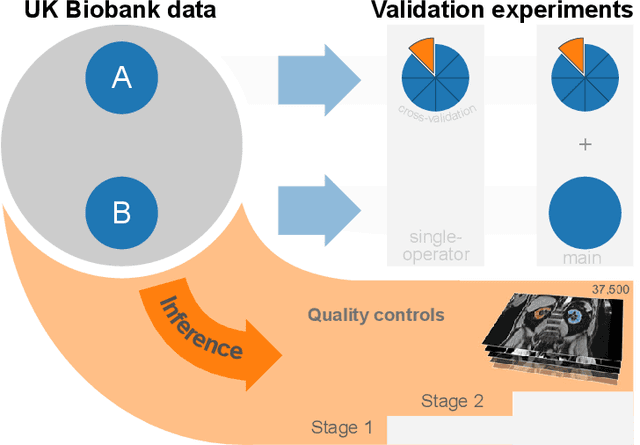
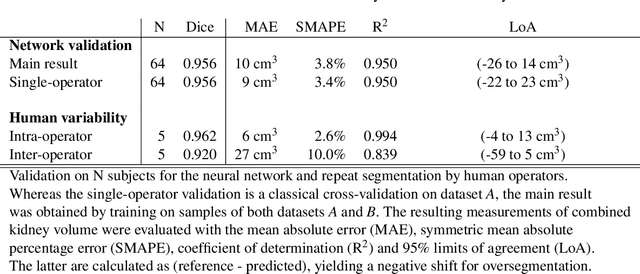
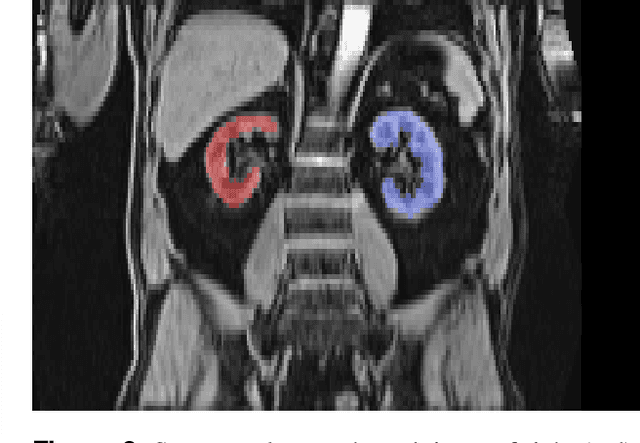
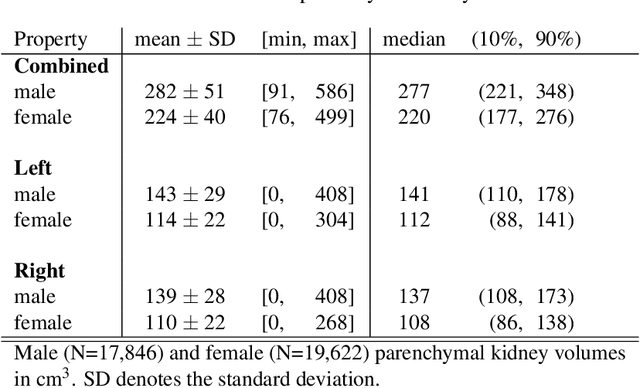
Abstract:The UK Biobank is collecting extensive data on health-related characteristics of over half a million volunteers. The biological samples of blood and urine can provide valuable insight on kidney function, with important links to cardiovascular and metabolic health. Further information on kidney anatomy could be obtained by medical imaging. In contrast to the brain, heart, liver, and pancreas, no dedicated Magnetic Resonance Imaging (MRI) is planned for the kidneys. An image-based assessment is nonetheless feasible in the neck-to-knee body MRI intended for abdominal body composition analysis, which also covers the kidneys. In this work, a pipeline for automated segmentation of parenchymal kidney volume in UK Biobank neck-to-knee body MRI is proposed. The underlying neural network reaches a relative error of 3.8%, with Dice score 0.956 in validation on 64 subjects, close to the 2.6% and Dice score 0.962 for repeated segmentation by one human operator. The released MRI of about 40,000 subjects can be processed within two days, yielding volume measurements of left and right kidney. Algorithmic quality ratings enabled the exclusion of outliers and potential failure cases. The resulting measurements can be studied and shared for large-scale investigation of associations and longitudinal changes in parenchymal kidney volume.
Fast Graph-Cut Based Optimization for Practical Dense Deformable Registration of Volume Images
Oct 19, 2018



Abstract:Objective: Deformable image registration is a fundamental problem in medical image analysis, with applications such as longitudinal studies, population modeling, and atlas based image segmentation. Registration is often phrased as an optimization problem, i.e., finding a deformation field that is optimal according to a given objective function. Discrete, combinatorial, optimization techniques have successfully been employed to solve the resulting optimization problem. Specifically, optimization based on $\alpha$-expansion with minimal graph cuts has been proposed as a powerful tool for image registration. The high computational cost of the graph-cut based optimization approach, however, limits the utility of this approach for registration of large volume images. Methods: Here, we propose to accelerate graph-cut based deformable registration by dividing the image into overlapping sub-regions and restricting the $\alpha$-expansion moves to a single sub-region at a time. Results: We demonstrate empirically that this approach can achieve a large reduction in computation time -- from days to minutes -- with only a small penalty in terms of solution quality. Conclusion: The reduction in computation time provided by the proposed method makes graph cut based deformable registration viable for large volume images. Significance: Graph cut based image registration has previously been shown to produce excellent results, but the high computational cost has hindered the adoption of the method for registration of large medical volume images. Our proposed method lifts this restriction, requiring only a small fraction of the computational cost to produce results of comparable quality.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge