Laurent Letourneau-Guillon
Analysis of the MICCAI Brain Tumor Segmentation -- Metastases (BraTS-METS) 2025 Lighthouse Challenge: Brain Metastasis Segmentation on Pre- and Post-treatment MRI
Apr 16, 2025Abstract:Despite continuous advancements in cancer treatment, brain metastatic disease remains a significant complication of primary cancer and is associated with an unfavorable prognosis. One approach for improving diagnosis, management, and outcomes is to implement algorithms based on artificial intelligence for the automated segmentation of both pre- and post-treatment MRI brain images. Such algorithms rely on volumetric criteria for lesion identification and treatment response assessment, which are still not available in clinical practice. Therefore, it is critical to establish tools for rapid volumetric segmentations methods that can be translated to clinical practice and that are trained on high quality annotated data. The BraTS-METS 2025 Lighthouse Challenge aims to address this critical need by establishing inter-rater and intra-rater variability in dataset annotation by generating high quality annotated datasets from four individual instances of segmentation by neuroradiologists while being recorded on video (two instances doing "from scratch" and two instances after AI pre-segmentation). This high-quality annotated dataset will be used for testing phase in 2025 Lighthouse challenge and will be publicly released at the completion of the challenge. The 2025 Lighthouse challenge will also release the 2023 and 2024 segmented datasets that were annotated using an established pipeline of pre-segmentation, student annotation, two neuroradiologists checking, and one neuroradiologist finalizing the process. It builds upon its previous edition by including post-treatment cases in the dataset. Using these high-quality annotated datasets, the 2025 Lighthouse challenge plans to test benchmark algorithms for automated segmentation of pre-and post-treatment brain metastases (BM), trained on diverse and multi-institutional datasets of MRI images obtained from patients with brain metastases.
Benchmarking the CoW with the TopCoW Challenge: Topology-Aware Anatomical Segmentation of the Circle of Willis for CTA and MRA
Dec 29, 2023



Abstract:The Circle of Willis (CoW) is an important network of arteries connecting major circulations of the brain. Its vascular architecture is believed to affect the risk, severity, and clinical outcome of serious neuro-vascular diseases. However, characterizing the highly variable CoW anatomy is still a manual and time-consuming expert task. The CoW is usually imaged by two angiographic imaging modalities, magnetic resonance angiography (MRA) and computed tomography angiography (CTA), but there exist limited public datasets with annotations on CoW anatomy, especially for CTA. Therefore we organized the TopCoW Challenge in 2023 with the release of an annotated CoW dataset and invited submissions worldwide for the CoW segmentation task, which attracted over 140 registered participants from four continents. TopCoW dataset was the first public dataset with voxel-level annotations for CoW's 13 vessel components, made possible by virtual-reality (VR) technology. It was also the first dataset with paired MRA and CTA from the same patients. TopCoW challenge aimed to tackle the CoW characterization problem as a multiclass anatomical segmentation task with an emphasis on topological metrics. The top performing teams managed to segment many CoW components to Dice scores around 90%, but with lower scores for communicating arteries and rare variants. There were also topological mistakes for predictions with high Dice scores. Additional topological analysis revealed further areas for improvement in detecting certain CoW components and matching CoW variant's topology accurately. TopCoW represented a first attempt at benchmarking the CoW anatomical segmentation task for MRA and CTA, both morphologically and topologically.
Beyond pixel-wise supervision for segmentation: A few global shape descriptors might be surprisingly good!
May 03, 2021

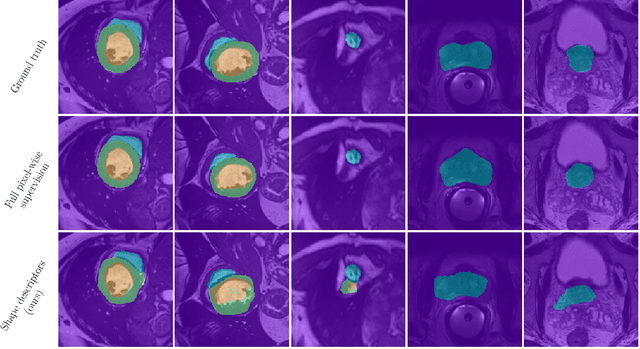

Abstract:Standard losses for training deep segmentation networks could be seen as individual classifications of pixels, instead of supervising the global shape of the predicted segmentations. While effective, they require exact knowledge of the label of each pixel in an image. This study investigates how effective global geometric shape descriptors could be, when used on their own as segmentation losses for training deep networks. Not only interesting theoretically, there exist deeper motivations to posing segmentation problems as a reconstruction of shape descriptors: Annotations to obtain approximations of low-order shape moments could be much less cumbersome than their full-mask counterparts, and anatomical priors could be readily encoded into invariant shape descriptions, which might alleviate the annotation burden. Also, and most importantly, we hypothesize that, given a task, certain shape descriptions might be invariant across image acquisition protocols/modalities and subject populations, which might open interesting research avenues for generalization in medical image segmentation. We introduce and formulate a few shape descriptors in the context of deep segmentation, and evaluate their potential as standalone losses on two different challenging tasks. Inspired by recent works in constrained optimization for deep networks, we propose a way to use those descriptors to supervise segmentation, without any pixel-level label. Very surprisingly, as little as 4 descriptors values per class can approach the performance of a segmentation mask with 65k individual discrete labels. We also found that shape descriptors can be a valid way to encode anatomical priors about the task, enabling to leverage expert knowledge without additional annotations. Our implementation is publicly available and can be easily extended to other tasks and descriptors: https://github.com/hkervadec/shape_descriptors
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge