Hervé Lombaert
Anatomically-aware conformal prediction for medical image segmentation with random walks
Jan 26, 2026Abstract:The reliable deployment of deep learning in medical imaging requires uncertainty quantification that provides rigorous error guarantees while remaining anatomically meaningful. Conformal prediction (CP) is a powerful distribution-free framework for constructing statistically valid prediction intervals. However, standard applications in segmentation often ignore anatomical context, resulting in fragmented, spatially incoherent, and over-segmented prediction sets that limit clinical utility. To bridge this gap, this paper proposes Random-Walk Conformal Prediction (RW-CP), a model-agnostic framework which can be added on top of any segmentation method. RW-CP enforces spatial coherence to generate anatomically valid sets. Our method constructs a k-nearest neighbour graph from pre-trained vision foundation model features and applies a random walk to diffuse uncertainty. The random walk diffusion regularizes the non-conformity scores, making the prediction sets less sensitive to the conformal calibration parameter $λ$, ensuring more stable and continuous anatomical boundaries. RW-CP maintains rigorous marginal coverage while significantly improving segmentation quality. Evaluations on multi-modal public datasets show improvements of up to $35.4\%$ compared to standard CP baselines, given an allowable error rate of $α=0.1$.
Prompt learning with bounding box constraints for medical image segmentation
Jul 03, 2025Abstract:Pixel-wise annotations are notoriously labourious and costly to obtain in the medical domain. To mitigate this burden, weakly supervised approaches based on bounding box annotations-much easier to acquire-offer a practical alternative. Vision foundation models have recently shown noteworthy segmentation performance when provided with prompts such as points or bounding boxes. Prompt learning exploits these models by adapting them to downstream tasks and automating segmentation, thereby reducing user intervention. However, existing prompt learning approaches depend on fully annotated segmentation masks. This paper proposes a novel framework that combines the representational power of foundation models with the annotation efficiency of weakly supervised segmentation. More specifically, our approach automates prompt generation for foundation models using only bounding box annotations. Our proposed optimization scheme integrates multiple constraints derived from box annotations with pseudo-labels generated by the prompted foundation model. Extensive experiments across multimodal datasets reveal that our weakly supervised method achieves an average Dice score of 84.90% in a limited data setting, outperforming existing fully-supervised and weakly-supervised approaches. The code is available at https://github.com/Minimel/box-prompt-learning-VFM.git
Automating MedSAM by Learning Prompts with Weak Few-Shot Supervision
Sep 30, 2024Abstract:Foundation models such as the recently introduced Segment Anything Model (SAM) have achieved remarkable results in image segmentation tasks. However, these models typically require user interaction through handcrafted prompts such as bounding boxes, which limits their deployment to downstream tasks. Adapting these models to a specific task with fully labeled data also demands expensive prior user interaction to obtain ground-truth annotations. This work proposes to replace conditioning on input prompts with a lightweight module that directly learns a prompt embedding from the image embedding, both of which are subsequently used by the foundation model to output a segmentation mask. Our foundation models with learnable prompts can automatically segment any specific region by 1) modifying the input through a prompt embedding predicted by a simple module, and 2) using weak labels (tight bounding boxes) and few-shot supervision (10 samples). Our approach is validated on MedSAM, a version of SAM fine-tuned for medical images, with results on three medical datasets in MR and ultrasound imaging. Our code is available on https://github.com/Minimel/MedSAMWeakFewShotPromptAutomation.
Neighbor-Aware Calibration of Segmentation Networks with Penalty-Based Constraints
Jan 25, 2024Abstract:Ensuring reliable confidence scores from deep neural networks is of paramount significance in critical decision-making systems, particularly in real-world domains such as healthcare. Recent literature on calibrating deep segmentation networks has resulted in substantial progress. Nevertheless, these approaches are strongly inspired by the advancements in classification tasks, and thus their uncertainty is usually modeled by leveraging the information of individual pixels, disregarding the local structure of the object of interest. Indeed, only the recent Spatially Varying Label Smoothing (SVLS) approach considers pixel spatial relationships across classes, by softening the pixel label assignments with a discrete spatial Gaussian kernel. In this work, we first present a constrained optimization perspective of SVLS and demonstrate that it enforces an implicit constraint on soft class proportions of surrounding pixels. Furthermore, our analysis shows that SVLS lacks a mechanism to balance the contribution of the constraint with the primary objective, potentially hindering the optimization process. Based on these observations, we propose NACL (Neighbor Aware CaLibration), a principled and simple solution based on equality constraints on the logit values, which enables to control explicitly both the enforced constraint and the weight of the penalty, offering more flexibility. Comprehensive experiments on a wide variety of well-known segmentation benchmarks demonstrate the superior calibration performance of the proposed approach, without affecting its discriminative power. Furthermore, ablation studies empirically show the model agnostic nature of our approach, which can be used to train a wide span of deep segmentation networks.
Trust your neighbours: Penalty-based constraints for model calibration
Mar 11, 2023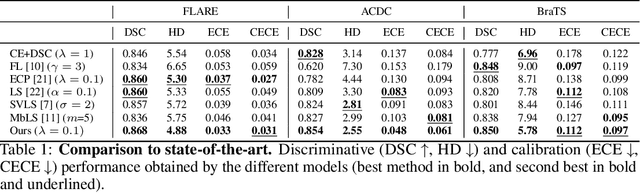
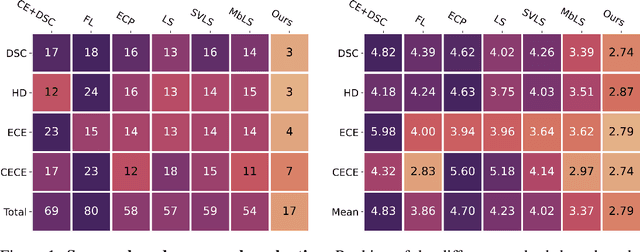
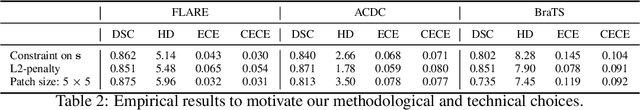
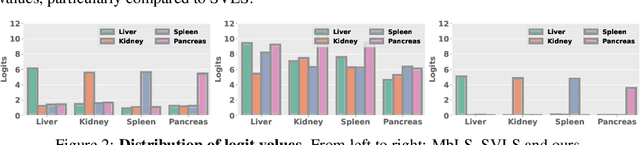
Abstract:Ensuring reliable confidence scores from deep networks is of pivotal importance in critical decision-making systems, notably in the medical domain. While recent literature on calibrating deep segmentation networks has led to significant progress, their uncertainty is usually modeled by leveraging the information of individual pixels, which disregards the local structure of the object of interest. In particular, only the recent Spatially Varying Label Smoothing (SVLS) approach addresses this issue by softening the pixel label assignments with a discrete spatial Gaussian kernel. In this work, we first present a constrained optimization perspective of SVLS and demonstrate that it enforces an implicit constraint on soft class proportions of surrounding pixels. Furthermore, our analysis shows that SVLS lacks a mechanism to balance the contribution of the constraint with the primary objective, potentially hindering the optimization process. Based on these observations, we propose a principled and simple solution based on equality constraints on the logit values, which enables to control explicitly both the enforced constraint and the weight of the penalty, offering more flexibility. Comprehensive experiments on a variety of well-known segmentation benchmarks demonstrate the superior performance of the proposed approach.
Active learning for medical image segmentation with stochastic batches
Jan 18, 2023



Abstract:The performance of learning-based algorithms improves with the amount of labelled data used for training. Yet, manually annotating data can be tedious and expensive, especially in medical image segmentation. To reduce manual labelling, active learning (AL) targets the most informative samples from the unlabelled set to annotate and add to the labelled training set. On one hand, most active learning works have focused on the classification or limited segmentation of natural images, despite active learning being highly desirable in the difficult task of medical image segmentation. On the other hand, uncertainty-based AL approaches notoriously offer sub-optimal batch-query strategies, while diversity-based methods tend to be computationally expensive. Over and above methodological hurdles, random sampling has proven an extremely difficult baseline to outperform when varying learning and sampling conditions. This work aims to take advantage of the diversity and speed offered by random sampling to improve the selection of uncertainty-based AL methods for segmenting medical images. More specifically, we propose to compute uncertainty at the level of batches instead of samples through an original use of stochastic batches during sampling in AL. Exhaustive experiments on medical image segmentation, with an illustration on MRI prostate imaging, show that the benefits of stochastic batches during sample selection are robust to a variety of changes in the training and sampling procedures.
TAAL: Test-time Augmentation for Active Learning in Medical Image Segmentation
Jan 16, 2023Abstract:Deep learning methods typically depend on the availability of labeled data, which is expensive and time-consuming to obtain. Active learning addresses such effort by prioritizing which samples are best to annotate in order to maximize the performance of the task model. While frameworks for active learning have been widely explored in the context of classification of natural images, they have been only sparsely used in medical image segmentation. The challenge resides in obtaining an uncertainty measure that reveals the best candidate data for annotation. This paper proposes Test-time Augmentation for Active Learning (TAAL), a novel semi-supervised active learning approach for segmentation that exploits the uncertainty information offered by data transformations. Our method applies cross-augmentation consistency during training and inference to both improve model learning in a semi-supervised fashion and identify the most relevant unlabeled samples to annotate next. In addition, our consistency loss uses a modified version of the JSD to further improve model performance. By relying on data transformations rather than on external modules or simple heuristics typically used in uncertainty-based strategies, TAAL emerges as a simple, yet powerful task-agnostic semi-supervised active learning approach applicable to the medical domain. Our results on a publicly-available dataset of cardiac images show that TAAL outperforms existing baseline methods in both fully-supervised and semi-supervised settings. Our implementation is publicly available on https://github.com/melinphd/TAAL.
Real-time simulation of viscoelastic tissue behavior with physics-guided deep learning
Jan 11, 2023Abstract:Finite element methods (FEM) are popular approaches for simulation of soft tissues with elastic or viscoelastic behavior. However, their usage in real-time applications, such as in virtual reality surgical training, is limited by computational cost. In this application scenario, which typically involves transportable simulators, the computing hardware severely constrains the size or the level of details of the simulated scene. To address this limitation, data-driven approaches have been suggested to simulate mechanical deformations by learning the mapping rules from FEM generated datasets. Herein, we propose a deep learning method for predicting displacement fields of soft tissues with viscoelastic properties. The main contribution of this work is the use of a physics-guided loss function for the optimization of the deep learning model parameters. The proposed deep learning model is based on convolutional (CNN) and recurrent layers (LSTM) to predict spatiotemporal variations. It is augmented with a mass conservation law in the lost function to prevent the generation of physically inconsistent results. The deep learning model is trained on a set of FEM datasets that are generated from a commercially available state-of-the-art numerical neurosurgery simulator. The use of the physics-guided loss function in a deep learning model has led to a better generalization in the prediction of deformations in unseen simulation cases. Moreover, the proposed method achieves a better accuracy over the conventional CNN models, where improvements were observed in unseen tissue from 8% to 30% depending on the magnitude of external forces. It is hoped that the present investigation will help in filling the gap in applying deep learning in virtual reality simulators, hence improving their computational performance (compared to FEM simulations) and ultimately their usefulness.
Test-Time Adaptation with Shape Moments for Image Segmentation
May 16, 2022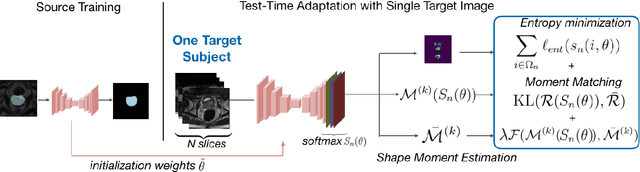
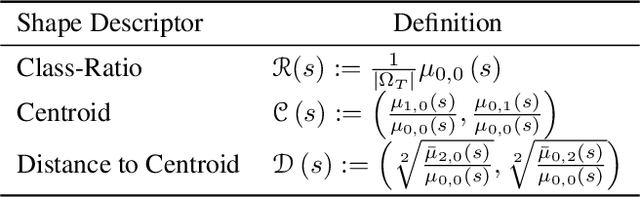
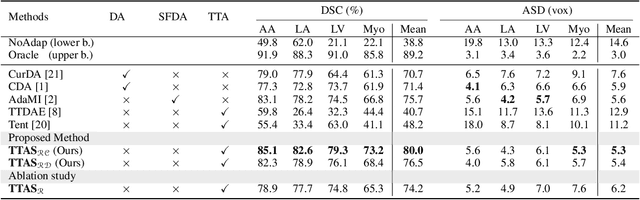

Abstract:Supervised learning is well-known to fail at generalization under distribution shifts. In typical clinical settings, the source data is inaccessible and the target distribution is represented with a handful of samples: adaptation can only happen at test time on a few or even a single subject(s). We investigate test-time single-subject adaptation for segmentation, and propose a Shape-guided Entropy Minimization objective for tackling this task. During inference for a single testing subject, our loss is minimized with respect to the batch normalization's scale and bias parameters. We show the potential of integrating various shape priors to guide adaptation to plausible solutions, and validate our method in two challenging scenarios: MRI-to-CT adaptation of cardiac segmentation and cross-site adaptation of prostate segmentation. Our approach exhibits substantially better performances than the existing test-time adaptation methods. Even more surprisingly, it fares better than state-of-the-art domain adaptation methods, although it forgoes training on additional target data during adaptation. Our results question the usefulness of training on target data in segmentation adaptation, and points to the substantial effect of shape priors on test-time inference. Our framework can be readily used for integrating various priors and for adapting any segmentation network, and our code is available.
Manifold-aware Synthesis of High-resolution Diffusion from Structural Imaging
Aug 11, 2021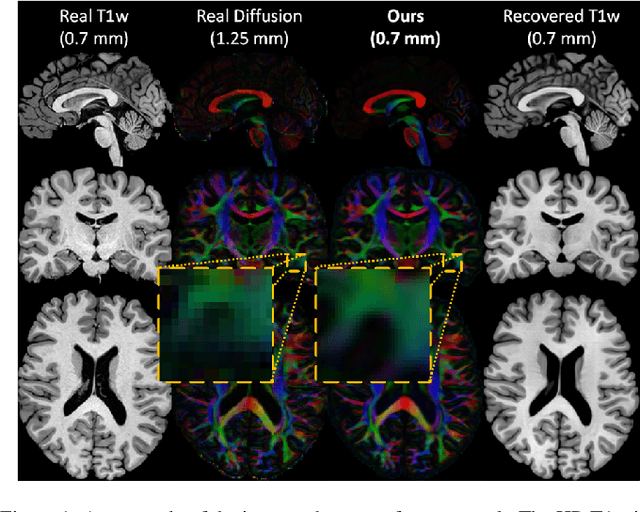
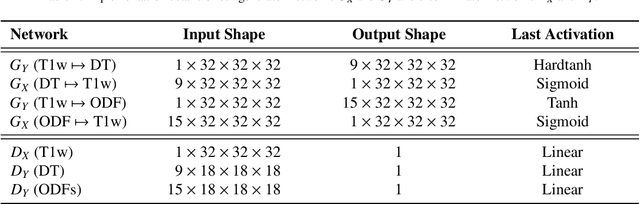
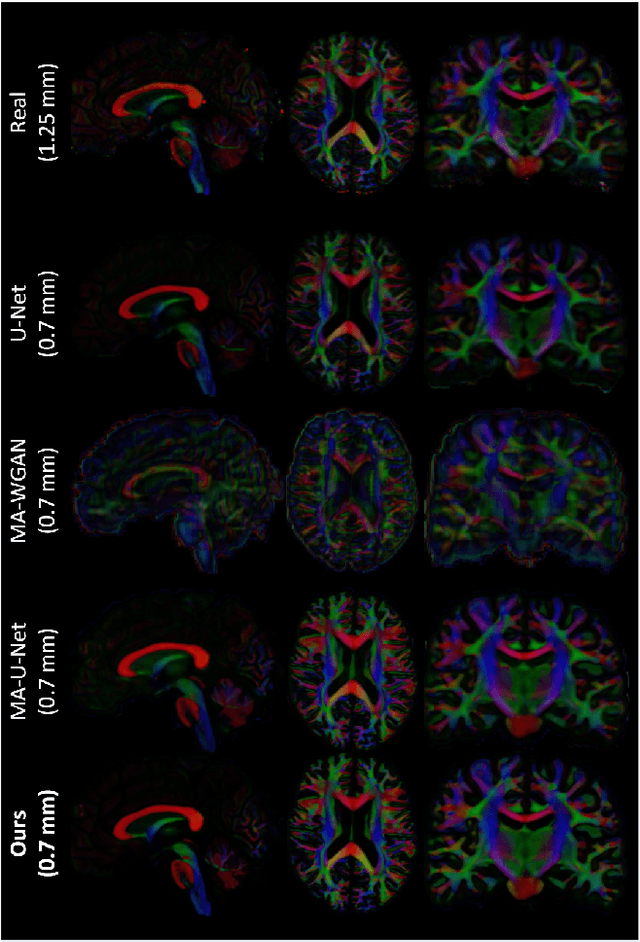
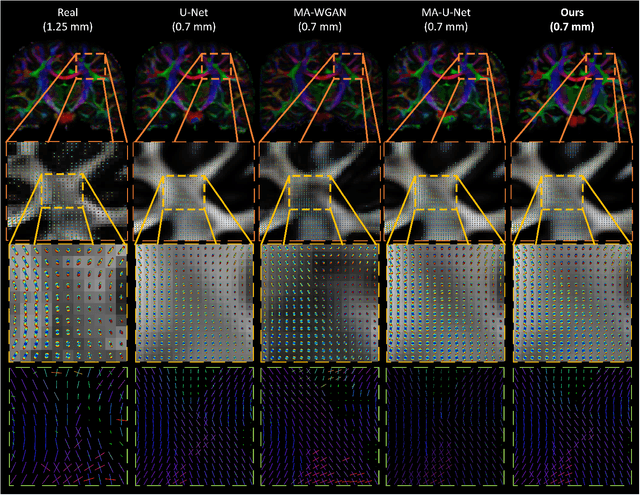
Abstract:The physical and clinical constraints surrounding diffusion-weighted imaging (DWI) often limit the spatial resolution of the produced images to voxels up to 8 times larger than those of T1w images. Thus, the detailed information contained in T1w imagescould help in the synthesis of diffusion images in higher resolution. However, the non-Euclidean nature of diffusion imaging hinders current deep generative models from synthesizing physically plausible images. In this work, we propose the first Riemannian network architecture for the direct generation of diffusion tensors (DT) and diffusion orientation distribution functions (dODFs) from high-resolution T1w images. Our integration of the Log-Euclidean Metric into a learning objective guarantees, unlike standard Euclidean networks, the mathematically-valid synthesis of diffusion. Furthermore, our approach improves the fractional anisotropy mean squared error (FA MSE) between the synthesized diffusion and the ground-truth by more than 23% and the cosine similarity between principal directions by almost 5% when compared to our baselines. We validate our generated diffusion by comparing the resulting tractograms to our expected real data. We observe similar fiber bundles with streamlines having less than 3% difference in length, less than 1% difference in volume, and a visually close shape. While our method is able to generate high-resolution diffusion images from structural inputs in less than 15 seconds, we acknowledge and discuss the limits of diffusion inference solely relying on T1w images. Our results nonetheless suggest a relationship between the high-level geometry of the brain and the overall white matter architecture.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge