Ao Chang
Subtyping Breast Lesions via Generative Augmentation based Long-tailed Recognition in Ultrasound
Jul 30, 2025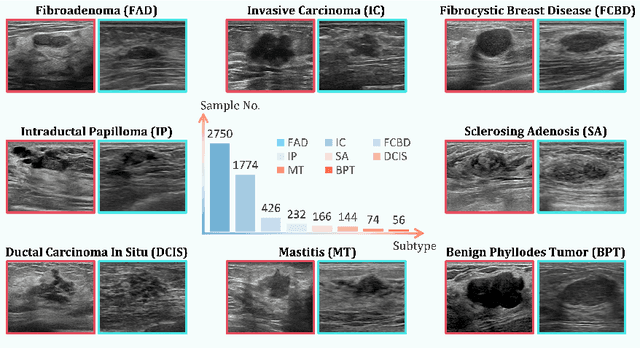
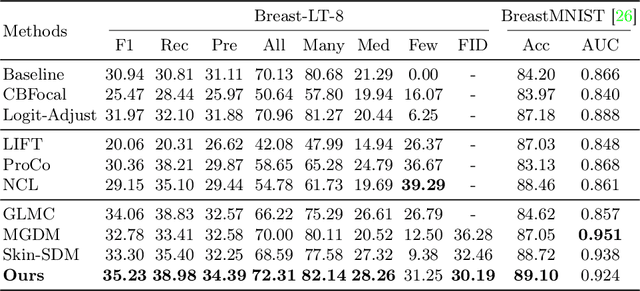
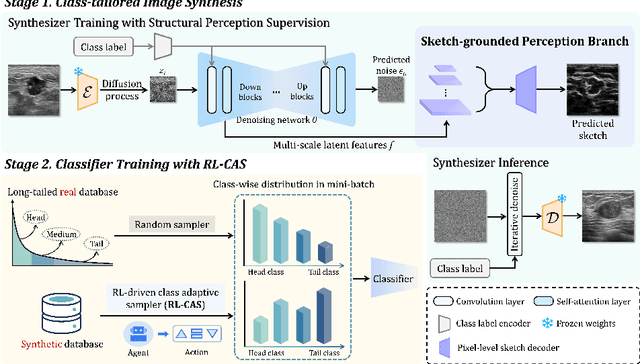
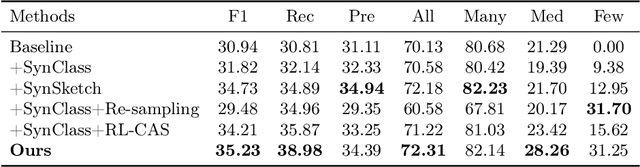
Abstract:Accurate identification of breast lesion subtypes can facilitate personalized treatment and interventions. Ultrasound (US), as a safe and accessible imaging modality, is extensively employed in breast abnormality screening and diagnosis. However, the incidence of different subtypes exhibits a skewed long-tailed distribution, posing significant challenges for automated recognition. Generative augmentation provides a promising solution to rectify data distribution. Inspired by this, we propose a dual-phase framework for long-tailed classification that mitigates distributional bias through high-fidelity data synthesis while avoiding overuse that corrupts holistic performance. The framework incorporates a reinforcement learning-driven adaptive sampler, dynamically calibrating synthetic-real data ratios by training a strategic multi-agent to compensate for scarcities of real data while ensuring stable discriminative capability. Furthermore, our class-controllable synthetic network integrates a sketch-grounded perception branch that harnesses anatomical priors to maintain distinctive class features while enabling annotation-free inference. Extensive experiments on an in-house long-tailed and a public imbalanced breast US datasets demonstrate that our method achieves promising performance compared to state-of-the-art approaches. More synthetic images can be found at https://github.com/Stinalalala/Breast-LT-GenAug.
Flip Learning: Weakly Supervised Erase to Segment Nodules in Breast Ultrasound
Mar 27, 2025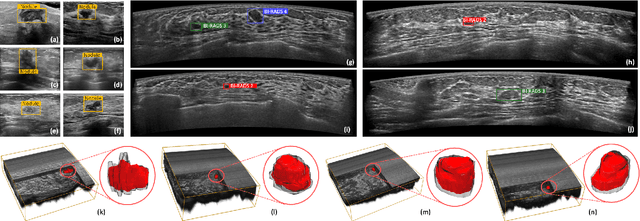
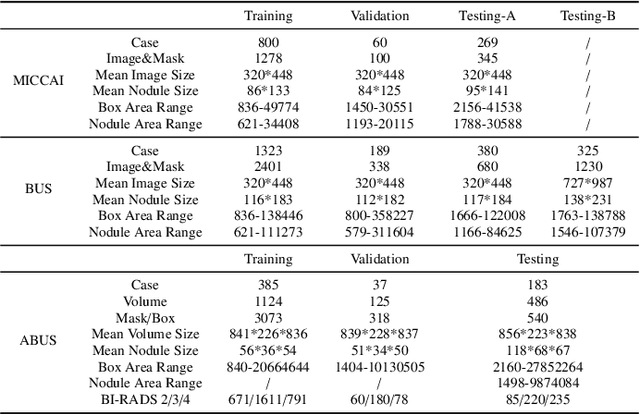
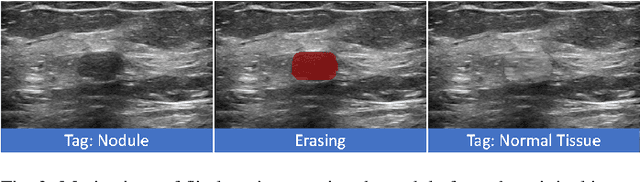
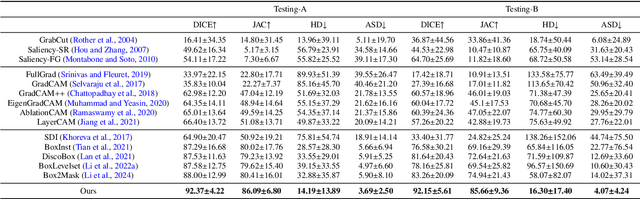
Abstract:Accurate segmentation of nodules in both 2D breast ultrasound (BUS) and 3D automated breast ultrasound (ABUS) is crucial for clinical diagnosis and treatment planning. Therefore, developing an automated system for nodule segmentation can enhance user independence and expedite clinical analysis. Unlike fully-supervised learning, weakly-supervised segmentation (WSS) can streamline the laborious and intricate annotation process. However, current WSS methods face challenges in achieving precise nodule segmentation, as many of them depend on inaccurate activation maps or inefficient pseudo-mask generation algorithms. In this study, we introduce a novel multi-agent reinforcement learning-based WSS framework called Flip Learning, which relies solely on 2D/3D boxes for accurate segmentation. Specifically, multiple agents are employed to erase the target from the box to facilitate classification tag flipping, with the erased region serving as the predicted segmentation mask. The key contributions of this research are as follows: (1) Adoption of a superpixel/supervoxel-based approach to encode the standardized environment, capturing boundary priors and expediting the learning process. (2) Introduction of three meticulously designed rewards, comprising a classification score reward and two intensity distribution rewards, to steer the agents' erasing process precisely, thereby avoiding both under- and over-segmentation. (3) Implementation of a progressive curriculum learning strategy to enable agents to interact with the environment in a progressively challenging manner, thereby enhancing learning efficiency. Extensively validated on the large in-house BUS and ABUS datasets, our Flip Learning method outperforms state-of-the-art WSS methods and foundation models, and achieves comparable performance as fully-supervised learning algorithms.
EM-Net: Efficient Channel and Frequency Learning with Mamba for 3D Medical Image Segmentation
Sep 26, 2024Abstract:Convolutional neural networks have primarily led 3D medical image segmentation but may be limited by small receptive fields. Transformer models excel in capturing global relationships through self-attention but are challenged by high computational costs at high resolutions. Recently, Mamba, a state space model, has emerged as an effective approach for sequential modeling. Inspired by its success, we introduce a novel Mamba-based 3D medical image segmentation model called EM-Net. It not only efficiently captures attentive interaction between regions by integrating and selecting channels, but also effectively utilizes frequency domain to harmonize the learning of features across varying scales, while accelerating training speed. Comprehensive experiments on two challenging multi-organ datasets with other state-of-the-art (SOTA) algorithms show that our method exhibits better segmentation accuracy while requiring nearly half the parameter size of SOTA models and 2x faster training speed.
Ctrl-GenAug: Controllable Generative Augmentation for Medical Sequence Classification
Sep 25, 2024



Abstract:In the medical field, the limited availability of large-scale datasets and labor-intensive annotation processes hinder the performance of deep models. Diffusion-based generative augmentation approaches present a promising solution to this issue, having been proven effective in advancing downstream medical recognition tasks. Nevertheless, existing works lack sufficient semantic and sequential steerability for challenging video/3D sequence generation, and neglect quality control of noisy synthesized samples, resulting in unreliable synthetic databases and severely limiting the performance of downstream tasks. In this work, we present Ctrl-GenAug, a novel and general generative augmentation framework that enables highly semantic- and sequential-customized sequence synthesis and suppresses incorrectly synthesized samples, to aid medical sequence classification. Specifically, we first design a multimodal conditions-guided sequence generator for controllably synthesizing diagnosis-promotive samples. A sequential augmentation module is integrated to enhance the temporal/stereoscopic coherence of generated samples. Then, we propose a noisy synthetic data filter to suppress unreliable cases at semantic and sequential levels. Extensive experiments on 3 medical datasets, using 11 networks trained on 3 paradigms, comprehensively analyze the effectiveness and generality of Ctrl-GenAug, particularly in underrepresented high-risk populations and out-domain conditions.
PE-MED: Prompt Enhancement for Interactive Medical Image Segmentation
Aug 26, 2023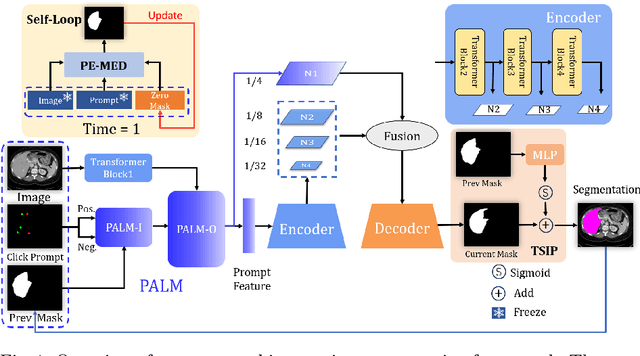
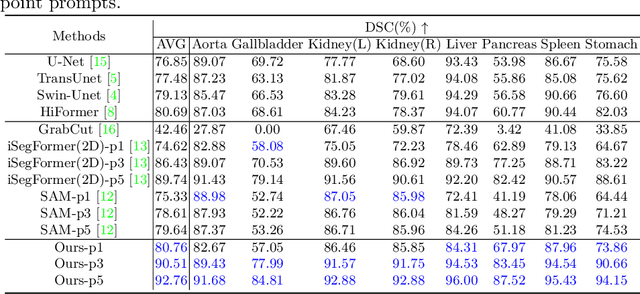
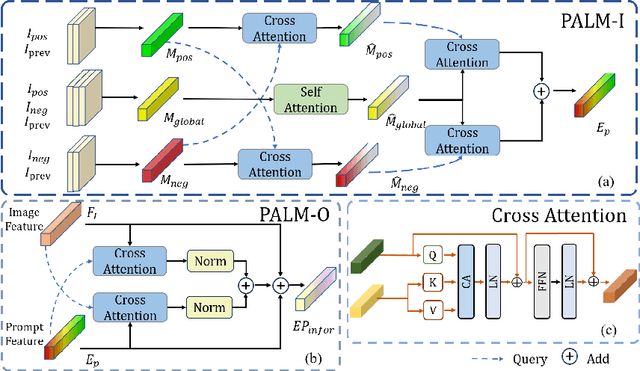
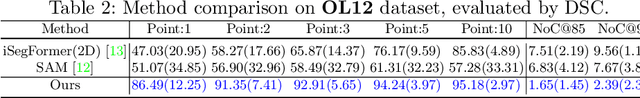
Abstract:Interactive medical image segmentation refers to the accurate segmentation of the target of interest through interaction (e.g., click) between the user and the image. It has been widely studied in recent years as it is less dependent on abundant annotated data and more flexible than fully automated segmentation. However, current studies have not fully explored user-provided prompt information (e.g., points), including the knowledge mined in one interaction, and the relationship between multiple interactions. Thus, in this paper, we introduce a novel framework equipped with prompt enhancement, called PE-MED, for interactive medical image segmentation. First, we introduce a Self-Loop strategy to generate warm initial segmentation results based on the first prompt. It can prevent the highly unfavorable scenarios, such as encountering a blank mask as the initial input after the first interaction. Second, we propose a novel Prompt Attention Learning Module (PALM) to mine useful prompt information in one interaction, enhancing the responsiveness of the network to user clicks. Last, we build a Time Series Information Propagation (TSIP) mechanism to extract the temporal relationships between multiple interactions and increase the model stability. Comparative experiments with other state-of-the-art (SOTA) medical image segmentation algorithms show that our method exhibits better segmentation accuracy and stability.
OnUVS: Online Feature Decoupling Framework for High-Fidelity Ultrasound Video Synthesis
Aug 16, 2023



Abstract:Ultrasound (US) imaging is indispensable in clinical practice. To diagnose certain diseases, sonographers must observe corresponding dynamic anatomic structures to gather comprehensive information. However, the limited availability of specific US video cases causes teaching difficulties in identifying corresponding diseases, which potentially impacts the detection rate of such cases. The synthesis of US videos may represent a promising solution to this issue. Nevertheless, it is challenging to accurately animate the intricate motion of dynamic anatomic structures while preserving image fidelity. To address this, we present a novel online feature-decoupling framework called OnUVS for high-fidelity US video synthesis. Our highlights can be summarized by four aspects. First, we introduced anatomic information into keypoint learning through a weakly-supervised training strategy, resulting in improved preservation of anatomical integrity and motion while minimizing the labeling burden. Second, to better preserve the integrity and textural information of US images, we implemented a dual-decoder that decouples the content and textural features in the generator. Third, we adopted a multiple-feature discriminator to extract a comprehensive range of visual cues, thereby enhancing the sharpness and fine details of the generated videos. Fourth, we constrained the motion trajectories of keypoints during online learning to enhance the fluidity of generated videos. Our validation and user studies on in-house echocardiographic and pelvic floor US videos showed that OnUVS synthesizes US videos with high fidelity.
Segment Anything Model for Medical Images?
May 01, 2023



Abstract:The Segment Anything Model (SAM) is the first foundation model for general image segmentation. It designed a novel promotable segmentation task, ensuring zero-shot image segmentation using the pre-trained model via two main modes including automatic everything and manual prompt. SAM has achieved impressive results on various natural image segmentation tasks. However, medical image segmentation (MIS) is more challenging due to the complex modalities, fine anatomical structures, uncertain and complex object boundaries, and wide-range object scales. Meanwhile, zero-shot and efficient MIS can well reduce the annotation time and boost the development of medical image analysis. Hence, SAM seems to be a potential tool and its performance on large medical datasets should be further validated. We collected and sorted 52 open-source datasets, and built a large medical segmentation dataset with 16 modalities, 68 objects, and 553K slices. We conducted a comprehensive analysis of different SAM testing strategies on the so-called COSMOS 553K dataset. Extensive experiments validate that SAM performs better with manual hints like points and boxes for object perception in medical images, leading to better performance in prompt mode compared to everything mode. Additionally, SAM shows remarkable performance in some specific objects and modalities, but is imperfect or even totally fails in other situations. Finally, we analyze the influence of different factors (e.g., the Fourier-based boundary complexity and size of the segmented objects) on SAM's segmentation performance. Extensive experiments validate that SAM's zero-shot segmentation capability is not sufficient to ensure its direct application to the MIS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge