Jiongquan Chen
Fine-grained Context and Multi-modal Alignment for Freehand 3D Ultrasound Reconstruction
Jul 05, 2024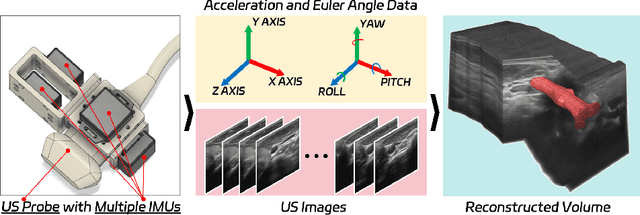


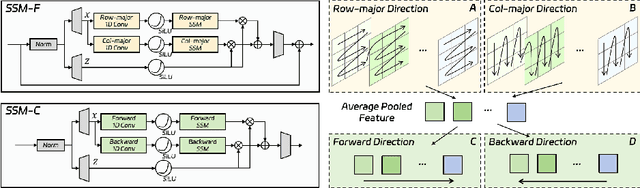
Abstract:Fine-grained spatio-temporal learning is crucial for freehand 3D ultrasound reconstruction. Previous works mainly resorted to the coarse-grained spatial features and the separated temporal dependency learning and struggles for fine-grained spatio-temporal learning. Mining spatio-temporal information in fine-grained scales is extremely challenging due to learning difficulties in long-range dependencies. In this context, we propose a novel method to exploit the long-range dependency management capabilities of the state space model (SSM) to address the above challenge. Our contribution is three-fold. First, we propose ReMamba, which mines multi-scale spatio-temporal information by devising a multi-directional SSM. Second, we propose an adaptive fusion strategy that introduces multiple inertial measurement units as auxiliary temporal information to enhance spatio-temporal perception. Last, we design an online alignment strategy that encodes the temporal information as pseudo labels for multi-modal alignment to further improve reconstruction performance. Extensive experimental validations on two large-scale datasets show remarkable improvement from our method over competitors.
A Foundation Model for General Moving Object Segmentation in Medical Images
Oct 04, 2023



Abstract:Medical image segmentation aims to delineate the anatomical or pathological structures of interest, playing a crucial role in clinical diagnosis. A substantial amount of high-quality annotated data is crucial for constructing high-precision deep segmentation models. However, medical annotation is highly cumbersome and time-consuming, especially for medical videos or 3D volumes, due to the huge labeling space and poor inter-frame consistency. Recently, a fundamental task named Moving Object Segmentation (MOS) has made significant advancements in natural images. Its objective is to delineate moving objects from the background within image sequences, requiring only minimal annotations. In this paper, we propose the first foundation model, named iMOS, for MOS in medical images. Extensive experiments on a large multi-modal medical dataset validate the effectiveness of the proposed iMOS. Specifically, with the annotation of only a small number of images in the sequence, iMOS can achieve satisfactory tracking and segmentation performance of moving objects throughout the entire sequence in bi-directions. We hope that the proposed iMOS can help accelerate the annotation speed of experts, and boost the development of medical foundation models.
Multi-IMU with Online Self-Consistency for Freehand 3D Ultrasound Reconstruction
Jul 19, 2023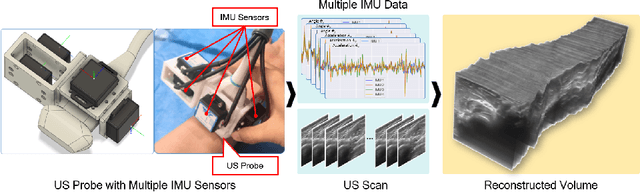
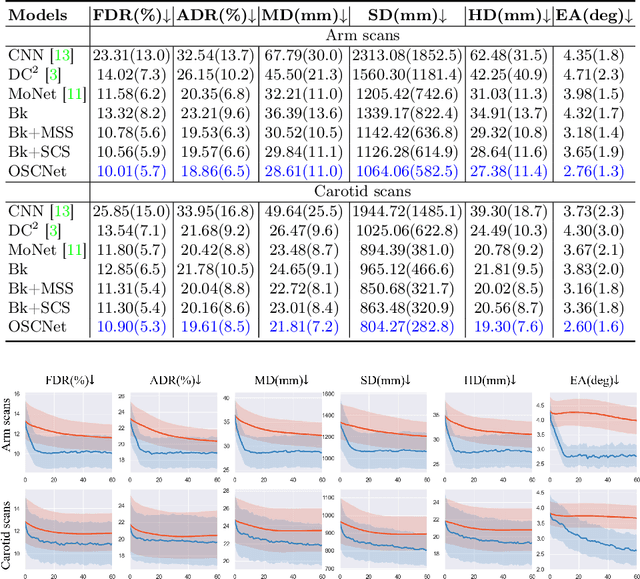
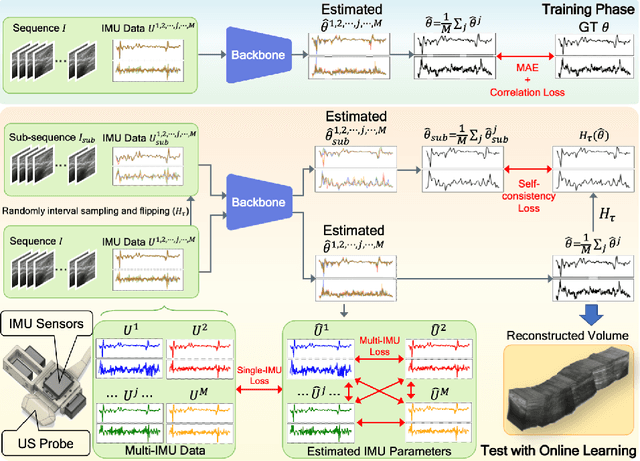
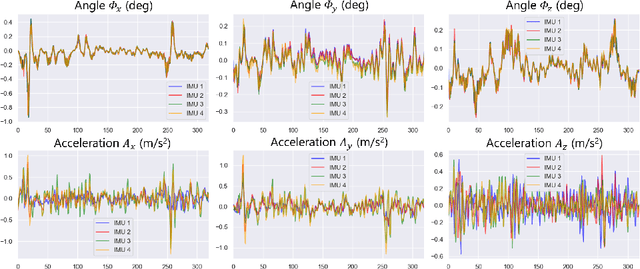
Abstract:Ultrasound (US) imaging is a popular tool in clinical diagnosis, offering safety, repeatability, and real-time capabilities. Freehand 3D US is a technique that provides a deeper understanding of scanned regions without increasing complexity. However, estimating elevation displacement and accumulation error remains challenging, making it difficult to infer the relative position using images alone. The addition of external lightweight sensors has been proposed to enhance reconstruction performance without adding complexity, which has been shown to be beneficial. We propose a novel online self-consistency network (OSCNet) using multiple inertial measurement units (IMUs) to improve reconstruction performance. OSCNet utilizes a modal-level self-supervised strategy to fuse multiple IMU information and reduce differences between reconstruction results obtained from each IMU data. Additionally, a sequence-level self-consistency strategy is proposed to improve the hierarchical consistency of prediction results among the scanning sequence and its sub-sequences. Experiments on large-scale arm and carotid datasets with multiple scanning tactics demonstrate that our OSCNet outperforms previous methods, achieving state-of-the-art reconstruction performance.
Instructive Feature Enhancement for Dichotomous Medical Image Segmentation
Jun 06, 2023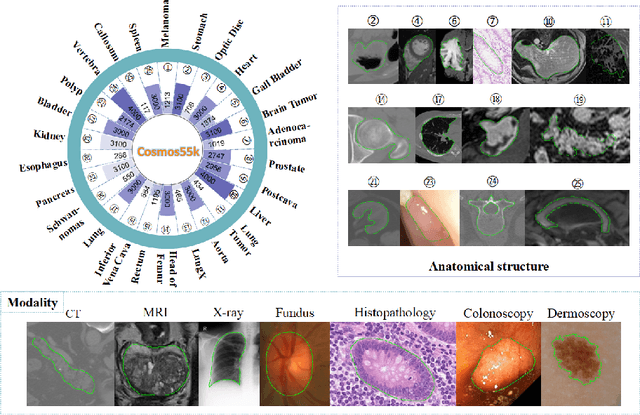

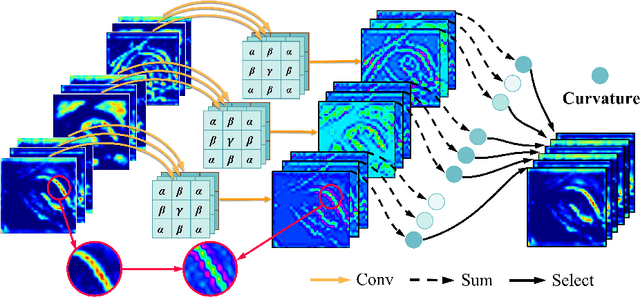
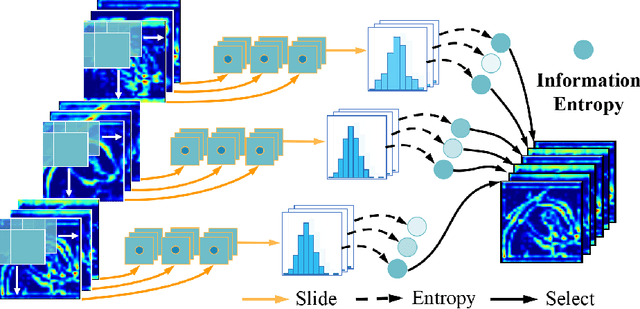
Abstract:Deep neural networks have been widely applied in dichotomous medical image segmentation (DMIS) of many anatomical structures in several modalities, achieving promising performance. However, existing networks tend to struggle with task-specific, heavy and complex designs to improve accuracy. They made little instructions to which feature channels would be more beneficial for segmentation, and that may be why the performance and universality of these segmentation models are hindered. In this study, we propose an instructive feature enhancement approach, namely IFE, to adaptively select feature channels with rich texture cues and strong discriminability to enhance raw features based on local curvature or global information entropy criteria. Being plug-and-play and applicable for diverse DMIS tasks, IFE encourages the model to focus on texture-rich features which are especially important for the ambiguous and challenging boundary identification, simultaneously achieving simplicity, universality, and certain interpretability. To evaluate the proposed IFE, we constructed the first large-scale DMIS dataset Cosmos55k, which contains 55,023 images from 7 modalities and 26 anatomical structures. Extensive experiments show that IFE can improve the performance of classic segmentation networks across different anatomies and modalities with only slight modifications. Code is available at https://github.com/yezi-66/IFE
Segment Anything Model for Medical Images?
May 01, 2023



Abstract:The Segment Anything Model (SAM) is the first foundation model for general image segmentation. It designed a novel promotable segmentation task, ensuring zero-shot image segmentation using the pre-trained model via two main modes including automatic everything and manual prompt. SAM has achieved impressive results on various natural image segmentation tasks. However, medical image segmentation (MIS) is more challenging due to the complex modalities, fine anatomical structures, uncertain and complex object boundaries, and wide-range object scales. Meanwhile, zero-shot and efficient MIS can well reduce the annotation time and boost the development of medical image analysis. Hence, SAM seems to be a potential tool and its performance on large medical datasets should be further validated. We collected and sorted 52 open-source datasets, and built a large medical segmentation dataset with 16 modalities, 68 objects, and 553K slices. We conducted a comprehensive analysis of different SAM testing strategies on the so-called COSMOS 553K dataset. Extensive experiments validate that SAM performs better with manual hints like points and boxes for object perception in medical images, leading to better performance in prompt mode compared to everything mode. Additionally, SAM shows remarkable performance in some specific objects and modalities, but is imperfect or even totally fails in other situations. Finally, we analyze the influence of different factors (e.g., the Fourier-based boundary complexity and size of the segmented objects) on SAM's segmentation performance. Extensive experiments validate that SAM's zero-shot segmentation capability is not sufficient to ensure its direct application to the MIS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge