Wenlong Shi
Few-shot Shape Recognition by Learning Deep Shape-aware Features
Dec 03, 2023Abstract:Traditional shape descriptors have been gradually replaced by convolutional neural networks due to their superior performance in feature extraction and classification. The state-of-the-art methods recognize object shapes via image reconstruction or pixel classification. However , these methods are biased toward texture information and overlook the essential shape descriptions, thus, they fail to generalize to unseen shapes. We are the first to propose a fewshot shape descriptor (FSSD) to recognize object shapes given only one or a few samples. We employ an embedding module for FSSD to extract transformation-invariant shape features. Secondly, we develop a dual attention mechanism to decompose and reconstruct the shape features via learnable shape primitives. In this way, any shape can be formed through a finite set basis, and the learned representation model is highly interpretable and extendable to unseen shapes. Thirdly, we propose a decoding module to include the supervision of shape masks and edges and align the original and reconstructed shape features, enforcing the learned features to be more shape-aware. Lastly, all the proposed modules are assembled into a few-shot shape recognition scheme. Experiments on five datasets show that our FSSD significantly improves the shape classification compared to the state-of-the-art under the few-shot setting.
FetusMapV2: Enhanced Fetal Pose Estimation in 3D Ultrasound
Oct 30, 2023Abstract:Fetal pose estimation in 3D ultrasound (US) involves identifying a set of associated fetal anatomical landmarks. Its primary objective is to provide comprehensive information about the fetus through landmark connections, thus benefiting various critical applications, such as biometric measurements, plane localization, and fetal movement monitoring. However, accurately estimating the 3D fetal pose in US volume has several challenges, including poor image quality, limited GPU memory for tackling high dimensional data, symmetrical or ambiguous anatomical structures, and considerable variations in fetal poses. In this study, we propose a novel 3D fetal pose estimation framework (called FetusMapV2) to overcome the above challenges. Our contribution is three-fold. First, we propose a heuristic scheme that explores the complementary network structure-unconstrained and activation-unreserved GPU memory management approaches, which can enlarge the input image resolution for better results under limited GPU memory. Second, we design a novel Pair Loss to mitigate confusion caused by symmetrical and similar anatomical structures. It separates the hidden classification task from the landmark localization task and thus progressively eases model learning. Last, we propose a shape priors-based self-supervised learning by selecting the relatively stable landmarks to refine the pose online. Extensive experiments and diverse applications on a large-scale fetal US dataset including 1000 volumes with 22 landmarks per volume demonstrate that our method outperforms other strong competitors.
A Foundation Model for General Moving Object Segmentation in Medical Images
Oct 04, 2023



Abstract:Medical image segmentation aims to delineate the anatomical or pathological structures of interest, playing a crucial role in clinical diagnosis. A substantial amount of high-quality annotated data is crucial for constructing high-precision deep segmentation models. However, medical annotation is highly cumbersome and time-consuming, especially for medical videos or 3D volumes, due to the huge labeling space and poor inter-frame consistency. Recently, a fundamental task named Moving Object Segmentation (MOS) has made significant advancements in natural images. Its objective is to delineate moving objects from the background within image sequences, requiring only minimal annotations. In this paper, we propose the first foundation model, named iMOS, for MOS in medical images. Extensive experiments on a large multi-modal medical dataset validate the effectiveness of the proposed iMOS. Specifically, with the annotation of only a small number of images in the sequence, iMOS can achieve satisfactory tracking and segmentation performance of moving objects throughout the entire sequence in bi-directions. We hope that the proposed iMOS can help accelerate the annotation speed of experts, and boost the development of medical foundation models.
Instructive Feature Enhancement for Dichotomous Medical Image Segmentation
Jun 06, 2023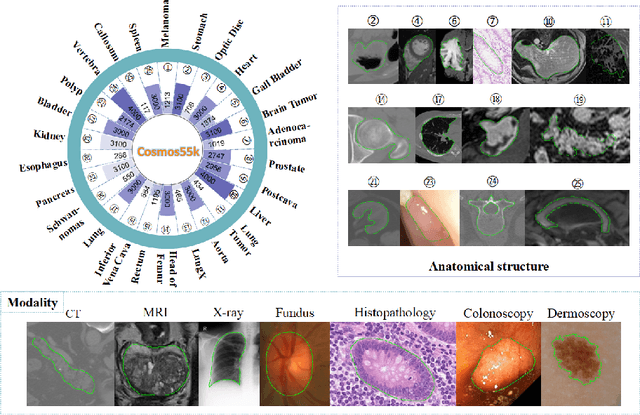

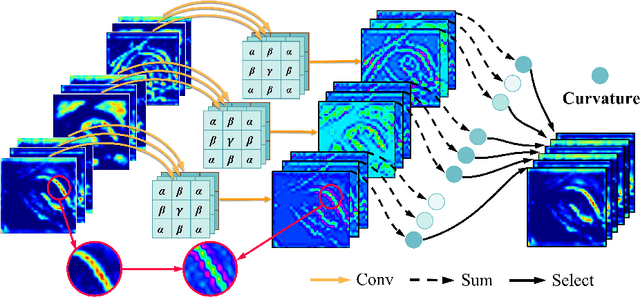
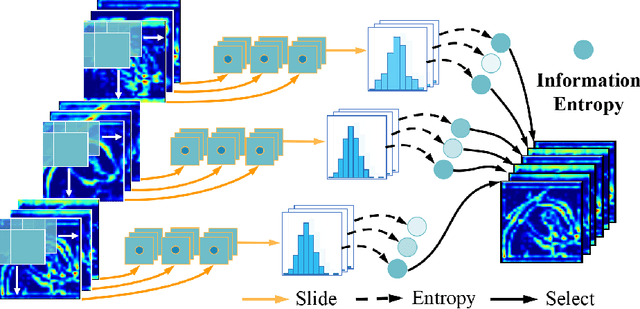
Abstract:Deep neural networks have been widely applied in dichotomous medical image segmentation (DMIS) of many anatomical structures in several modalities, achieving promising performance. However, existing networks tend to struggle with task-specific, heavy and complex designs to improve accuracy. They made little instructions to which feature channels would be more beneficial for segmentation, and that may be why the performance and universality of these segmentation models are hindered. In this study, we propose an instructive feature enhancement approach, namely IFE, to adaptively select feature channels with rich texture cues and strong discriminability to enhance raw features based on local curvature or global information entropy criteria. Being plug-and-play and applicable for diverse DMIS tasks, IFE encourages the model to focus on texture-rich features which are especially important for the ambiguous and challenging boundary identification, simultaneously achieving simplicity, universality, and certain interpretability. To evaluate the proposed IFE, we constructed the first large-scale DMIS dataset Cosmos55k, which contains 55,023 images from 7 modalities and 26 anatomical structures. Extensive experiments show that IFE can improve the performance of classic segmentation networks across different anatomies and modalities with only slight modifications. Code is available at https://github.com/yezi-66/IFE
Searching Collaborative Agents for Multi-plane Localization in 3D Ultrasound
May 22, 2021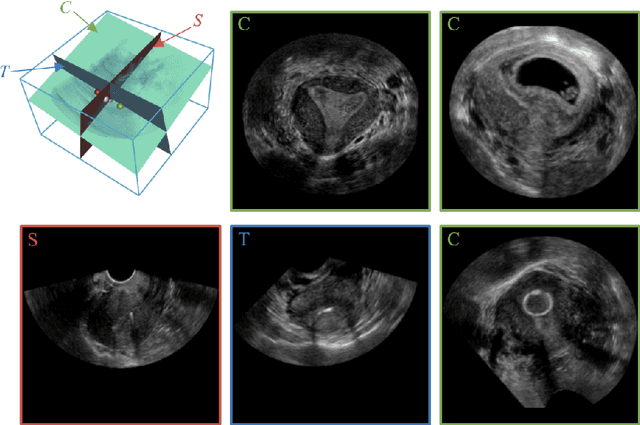
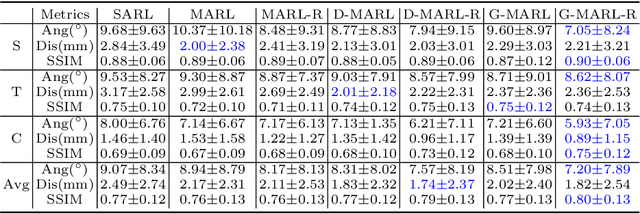
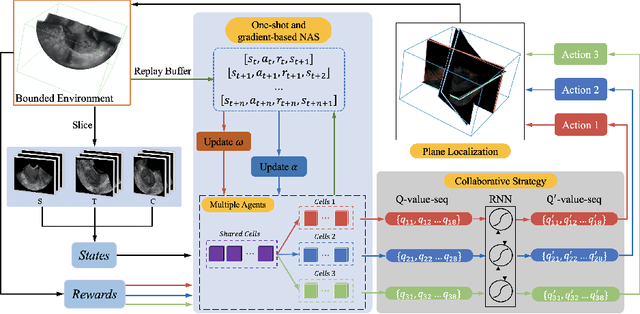

Abstract:3D ultrasound (US) has become prevalent due to its rich spatial and diagnostic information not contained in 2D US. Moreover, 3D US can contain multiple standard planes (SPs) in one shot. Thus, automatically localizing SPs in 3D US has the potential to improve user-independence and scanning-efficiency. However, manual SP localization in 3D US is challenging because of the low image quality, huge search space and large anatomical variability. In this work, we propose a novel multi-agent reinforcement learning (MARL) framework to simultaneously localize multiple SPs in 3D US. Our contribution is four-fold. First, our proposed method is general and it can accurately localize multiple SPs in different challenging US datasets. Second, we equip the MARL system with a recurrent neural network (RNN) based collaborative module, which can strengthen the communication among agents and learn the spatial relationship among planes effectively. Third, we explore to adopt the neural architecture search (NAS) to automatically design the network architecture of both the agents and the collaborative module. Last, we believe we are the first to realize automatic SP localization in pelvic US volumes, and note that our approach can handle both normal and abnormal uterus cases. Extensively validated on two challenging datasets of the uterus and fetal brain, our proposed method achieves the average localization accuracy of 7.03 degrees/1.59mm and 9.75 degrees/1.19mm. Experimental results show that our light-weight MARL model has higher accuracy than state-of-the-art methods.
Learn Fine-grained Adaptive Loss for Multiple Anatomical Landmark Detection in Medical Images
May 19, 2021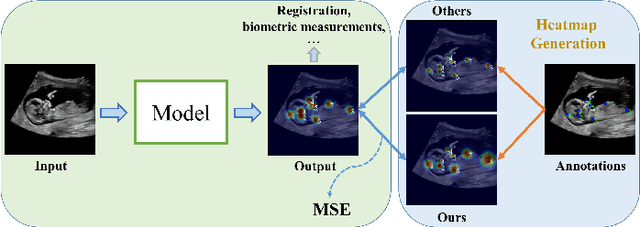
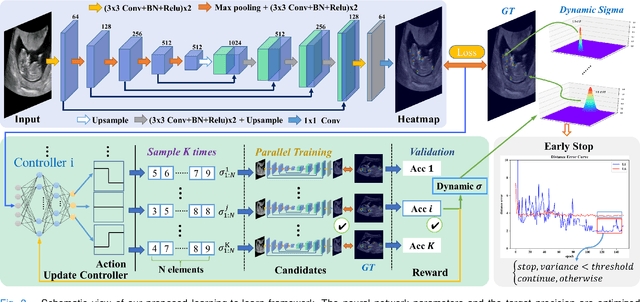
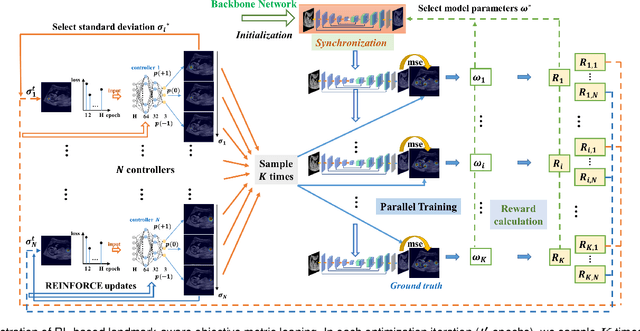
Abstract:Automatic and accurate detection of anatomical landmarks is an essential operation in medical image analysis with a multitude of applications. Recent deep learning methods have improved results by directly encoding the appearance of the captured anatomy with the likelihood maps (i.e., heatmaps). However, most current solutions overlook another essence of heatmap regression, the objective metric for regressing target heatmaps and rely on hand-crafted heuristics to set the target precision, thus being usually cumbersome and task-specific. In this paper, we propose a novel learning-to-learn framework for landmark detection to optimize the neural network and the target precision simultaneously. The pivot of this work is to leverage the reinforcement learning (RL) framework to search objective metrics for regressing multiple heatmaps dynamically during the training process, thus avoiding setting problem-specific target precision. We also introduce an early-stop strategy for active termination of the RL agent's interaction that adapts the optimal precision for separate targets considering exploration-exploitation tradeoffs. This approach shows better stability in training and improved localization accuracy in inference. Extensive experimental results on two different applications of landmark localization: 1) our in-house prenatal ultrasound (US) dataset and 2) the publicly available dataset of cephalometric X-Ray landmark detection, demonstrate the effectiveness of our proposed method. Our proposed framework is general and shows the potential to improve the efficiency of anatomical landmark detection.
Contrastive Rendering for Ultrasound Image Segmentation
Oct 10, 2020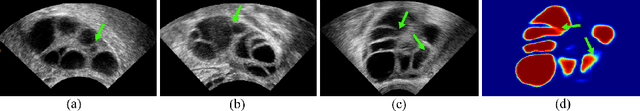
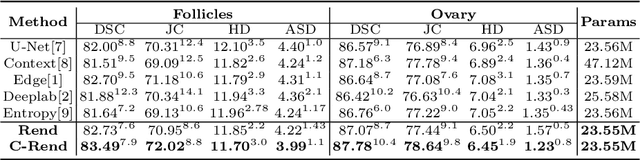
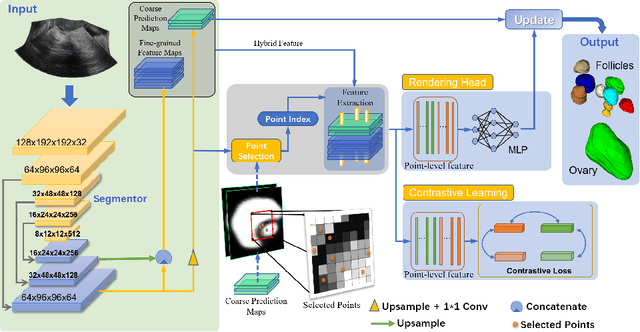
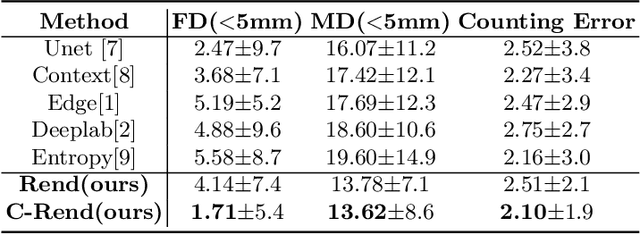
Abstract:Ultrasound (US) image segmentation embraced its significant improvement in deep learning era. However, the lack of sharp boundaries in US images still remains an inherent challenge for segmentation. Previous methods often resort to global context, multi-scale cues or auxiliary guidance to estimate the boundaries. It is hard for these methods to approach pixel-level learning for fine-grained boundary generating. In this paper, we propose a novel and effective framework to improve boundary estimation in US images. Our work has three highlights. First, we propose to formulate the boundary estimation as a rendering task, which can recognize ambiguous points (pixels/voxels) and calibrate the boundary prediction via enriched feature representation learning. Second, we introduce point-wise contrastive learning to enhance the similarity of points from the same class and contrastively decrease the similarity of points from different classes. Boundary ambiguities are therefore further addressed. Third, both rendering and contrastive learning tasks contribute to consistent improvement while reducing network parameters. As a proof-of-concept, we performed validation experiments on a challenging dataset of 86 ovarian US volumes. Results show that our proposed method outperforms state-of-the-art methods and has the potential to be used in clinical practice.
Region Proposal Network with Graph Prior and IoU-Balance Loss for Landmark Detection in 3D Ultrasound
Apr 01, 2020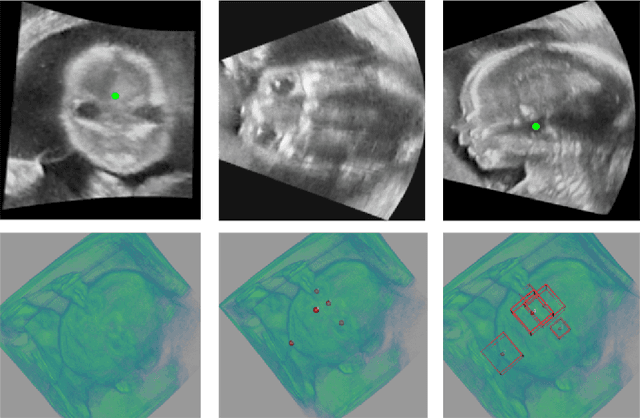
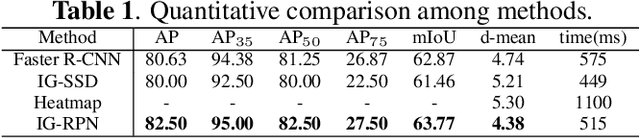
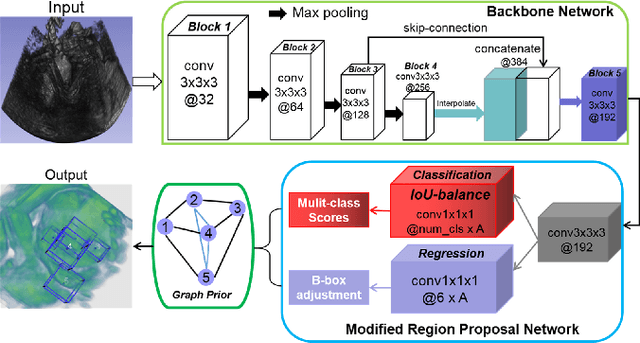
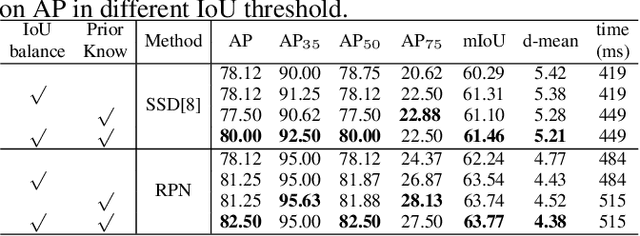
Abstract:3D ultrasound (US) can facilitate detailed prenatal examinations for fetal growth monitoring. To analyze a 3D US volume, it is fundamental to identify anatomical landmarks of the evaluated organs accurately. Typical deep learning methods usually regress the coordinates directly or involve heatmap-matching. However, these methods struggle to deal with volumes with large sizes and the highly-varying positions and orientations of fetuses. In this work, we exploit an object detection framework to detect landmarks in 3D fetal facial US volumes. By regressing multiple parameters of the landmark-centered bounding box (B-box) with a strict criteria, the proposed model is able to pinpoint the exact location of the targeted landmarks. Specifically, the model uses a 3D region proposal network (RPN) to generate 3D candidate regions, followed by several 3D classification branches to select the best candidate. It also adopts an IoU-balance loss to improve communications between branches that benefits the learning process. Furthermore, it leverages a distance-based graph prior to regularize the training and helps to reduce false positive predictions. The performance of the proposed framework is evaluated on a 3D US dataset to detect five key fetal facial landmarks. Results showed the proposed method outperforms some of the state-of-the-art methods in efficacy and efficiency.
FetusMap: Fetal Pose Estimation in 3D Ultrasound
Oct 11, 2019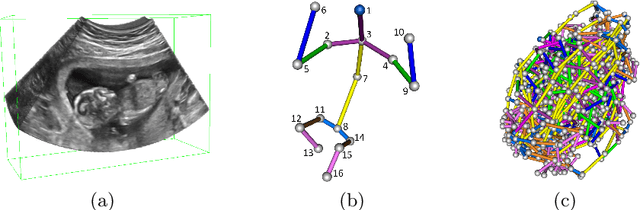
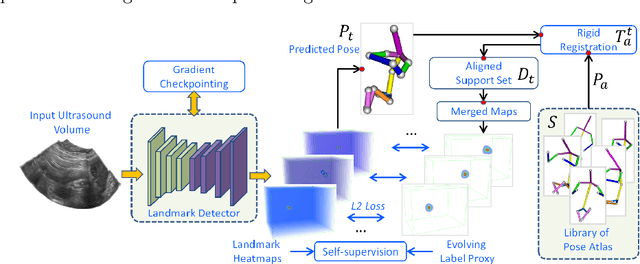
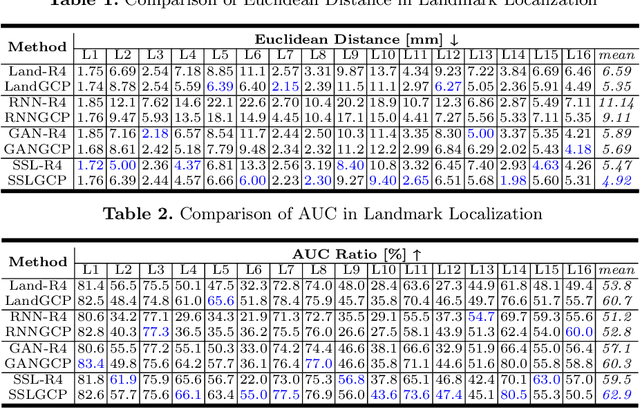
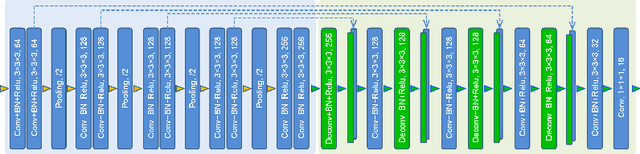
Abstract:The 3D ultrasound (US) entrance inspires a multitude of automated prenatal examinations. However, studies about the structuralized description of the whole fetus in 3D US are still rare. In this paper, we propose to estimate the 3D pose of fetus in US volumes to facilitate its quantitative analyses in global and local scales. Given the great challenges in 3D US, including the high volume dimension, poor image quality, symmetric ambiguity in anatomical structures and large variations of fetal pose, our contribution is three-fold. (i) This is the first work about 3D pose estimation of fetus in the literature. We aim to extract the skeleton of whole fetus and assign different segments/joints with correct torso/limb labels. (ii) We propose a self-supervised learning (SSL) framework to finetune the deep network to form visually plausible pose predictions. Specifically, we leverage the landmark-based registration to effectively encode case-adaptive anatomical priors and generate evolving label proxy for supervision. (iii) To enable our 3D network perceive better contextual cues with higher resolution input under limited computing resource, we further adopt the gradient check-pointing (GCP) strategy to save GPU memory and improve the prediction. Extensively validated on a large 3D US dataset, our method tackles varying fetal poses and achieves promising results. 3D pose estimation of fetus has potentials in serving as a map to provide navigation for many advanced studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge