Huanjia Luo
Weakly-supervised High-fidelity Ultrasound Video Synthesis with Feature Decoupling
Jul 01, 2022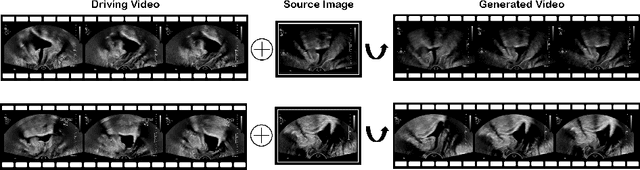
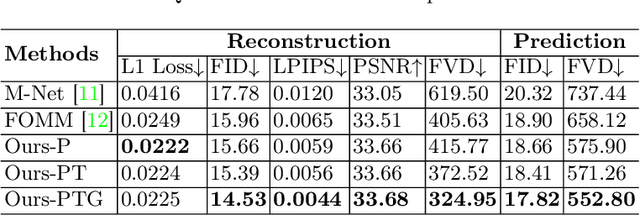
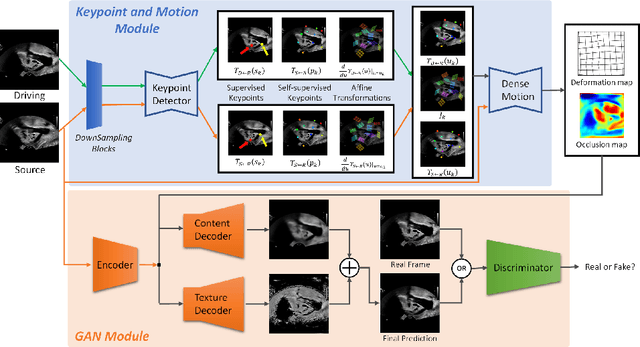
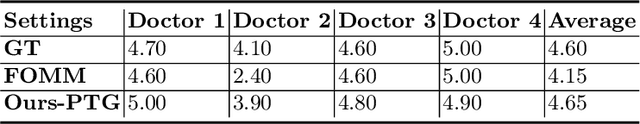
Abstract:Ultrasound (US) is widely used for its advantages of real-time imaging, radiation-free and portability. In clinical practice, analysis and diagnosis often rely on US sequences rather than a single image to obtain dynamic anatomical information. This is challenging for novices to learn because practicing with adequate videos from patients is clinically unpractical. In this paper, we propose a novel framework to synthesize high-fidelity US videos. Specifically, the synthesis videos are generated by animating source content images based on the motion of given driving videos. Our highlights are three-fold. First, leveraging the advantages of self- and fully-supervised learning, our proposed system is trained in weakly-supervised manner for keypoint detection. These keypoints then provide vital information for handling complex high dynamic motions in US videos. Second, we decouple content and texture learning using the dual decoders to effectively reduce the model learning difficulty. Last, we adopt the adversarial training strategy with GAN losses for further improving the sharpness of the generated videos, narrowing the gap between real and synthesis videos. We validate our method on a large in-house pelvic dataset with high dynamic motion. Extensive evaluation metrics and user study prove the effectiveness of our proposed method.
Agent with Warm Start and Adaptive Dynamic Termination for Plane Localization in 3D Ultrasound
Mar 26, 2021
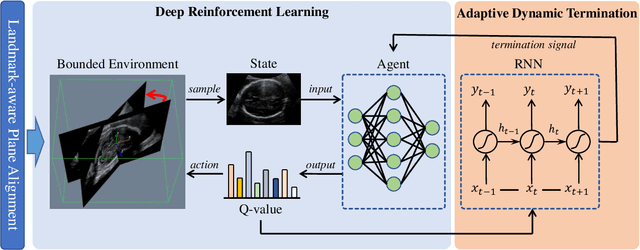
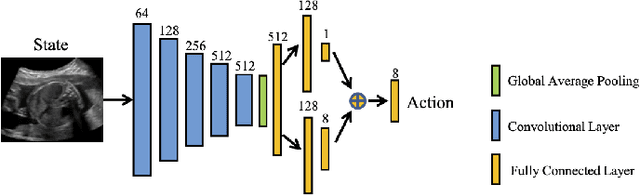
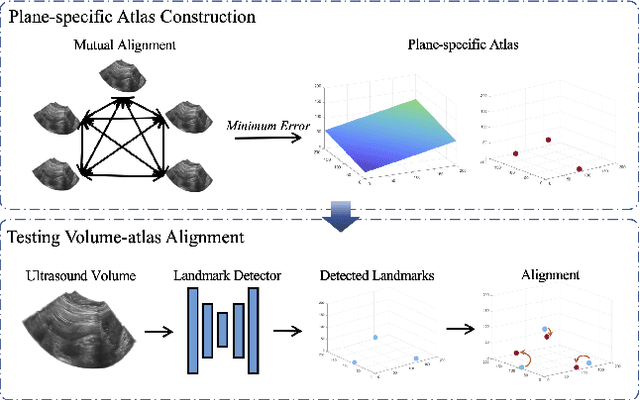
Abstract:Accurate standard plane (SP) localization is the fundamental step for prenatal ultrasound (US) diagnosis. Typically, dozens of US SPs are collected to determine the clinical diagnosis. 2D US has to perform scanning for each SP, which is time-consuming and operator-dependent. While 3D US containing multiple SPs in one shot has the inherent advantages of less user-dependency and more efficiency. Automatically locating SP in 3D US is very challenging due to the huge search space and large fetal posture variations. Our previous study proposed a deep reinforcement learning (RL) framework with an alignment module and active termination to localize SPs in 3D US automatically. However, termination of agent search in RL is important and affects the practical deployment. In this study, we enhance our previous RL framework with a newly designed adaptive dynamic termination to enable an early stop for the agent searching, saving at most 67% inference time, thus boosting the accuracy and efficiency of the RL framework at the same time. Besides, we validate the effectiveness and generalizability of our algorithm extensively on our in-house multi-organ datasets containing 433 fetal brain volumes, 519 fetal abdomen volumes, and 683 uterus volumes. Our approach achieves localization error of 2.52mm/10.26 degrees, 2.48mm/10.39 degrees, 2.02mm/10.48 degrees, 2.00mm/14.57 degrees, 2.61mm/9.71 degrees, 3.09mm/9.58 degrees, 1.49mm/7.54 degrees for the transcerebellar, transventricular, transthalamic planes in fetal brain, abdominal plane in fetal abdomen, and mid-sagittal, transverse and coronal planes in uterus, respectively. Experimental results show that our method is general and has the potential to improve the efficiency and standardization of US scanning.
Searching Collaborative Agents for Multi-plane Localization in 3D Ultrasound
Jul 30, 2020
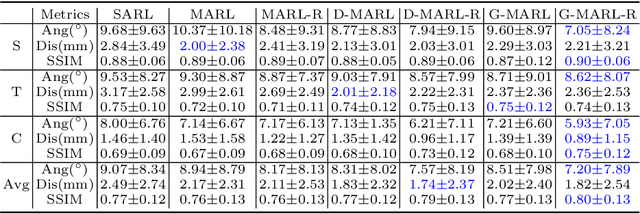
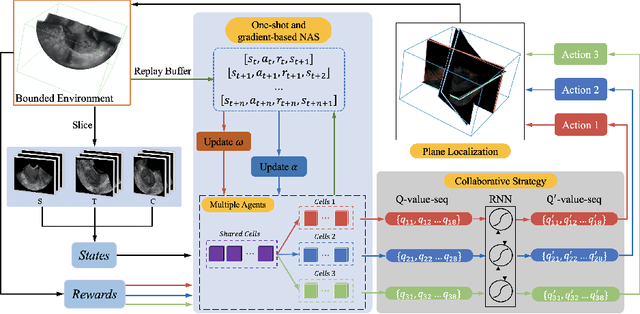

Abstract:3D ultrasound (US) is widely used due to its rich diagnostic information, portability and low cost. Automated standard plane (SP) localization in US volume not only improves efficiency and reduces user-dependence, but also boosts 3D US interpretation. In this study, we propose a novel Multi-Agent Reinforcement Learning (MARL) framework to localize multiple uterine SPs in 3D US simultaneously. Our contribution is two-fold. First, we equip the MARL with a one-shot neural architecture search (NAS) module to obtain the optimal agent for each plane. Specifically, Gradient-based search using Differentiable Architecture Sampler (GDAS) is employed to accelerate and stabilize the training process. Second, we propose a novel collaborative strategy to strengthen agents' communication. Our strategy uses recurrent neural network (RNN) to learn the spatial relationship among SPs effectively. Extensively validated on a large dataset, our approach achieves the accuracy of 7.05 degree/2.21mm, 8.62 degree/2.36mm and 5.93 degree/0.89mm for the mid-sagittal, transverse and coronal plane localization, respectively. The proposed MARL framework can significantly increase the plane localization accuracy and reduce the computational cost and model size.
Region Proposal Network with Graph Prior and IoU-Balance Loss for Landmark Detection in 3D Ultrasound
Apr 01, 2020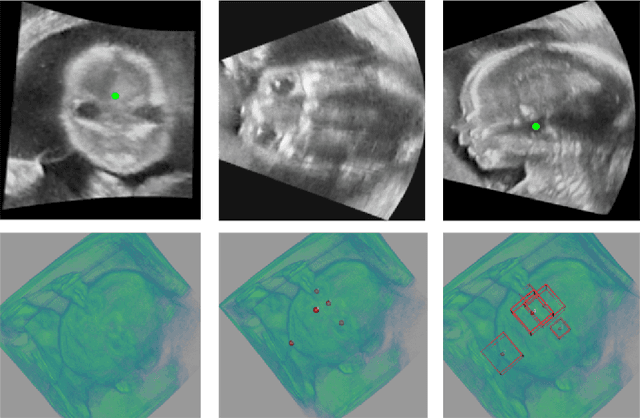
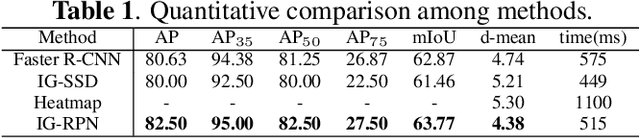
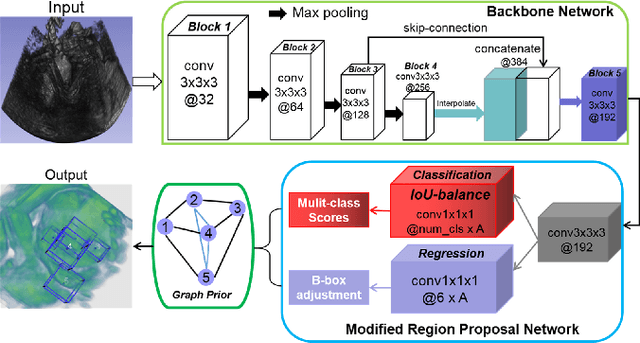
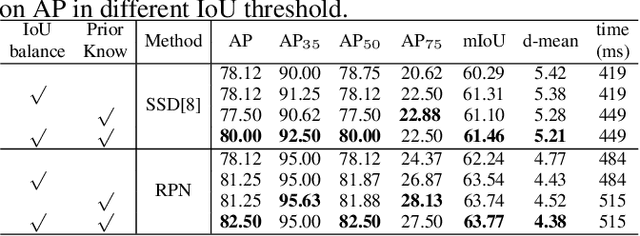
Abstract:3D ultrasound (US) can facilitate detailed prenatal examinations for fetal growth monitoring. To analyze a 3D US volume, it is fundamental to identify anatomical landmarks of the evaluated organs accurately. Typical deep learning methods usually regress the coordinates directly or involve heatmap-matching. However, these methods struggle to deal with volumes with large sizes and the highly-varying positions and orientations of fetuses. In this work, we exploit an object detection framework to detect landmarks in 3D fetal facial US volumes. By regressing multiple parameters of the landmark-centered bounding box (B-box) with a strict criteria, the proposed model is able to pinpoint the exact location of the targeted landmarks. Specifically, the model uses a 3D region proposal network (RPN) to generate 3D candidate regions, followed by several 3D classification branches to select the best candidate. It also adopts an IoU-balance loss to improve communications between branches that benefits the learning process. Furthermore, it leverages a distance-based graph prior to regularize the training and helps to reduce false positive predictions. The performance of the proposed framework is evaluated on a 3D US dataset to detect five key fetal facial landmarks. Results showed the proposed method outperforms some of the state-of-the-art methods in efficacy and efficiency.
Remove Appearance Shift for Ultrasound Image Segmentation via Fast and Universal Style Transfer
Feb 14, 2020



Abstract:Deep Neural Networks (DNNs) suffer from the performance degradation when image appearance shift occurs, especially in ultrasound (US) image segmentation. In this paper, we propose a novel and intuitive framework to remove the appearance shift, and hence improve the generalization ability of DNNs. Our work has three highlights. First, we follow the spirit of universal style transfer to remove appearance shifts, which was not explored before for US images. Without sacrificing image structure details, it enables the arbitrary style-content transfer. Second, accelerated with Adaptive Instance Normalization block, our framework achieved real-time speed required in the clinical US scanning. Third, an efficient and effective style image selection strategy is proposed to ensure the target-style US image and testing content US image properly match each other. Experiments on two large US datasets demonstrate that our methods are superior to state-of-the-art methods on making DNNs robust against various appearance shifts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge