Mingyuan Luo
Fine-grained Context and Multi-modal Alignment for Freehand 3D Ultrasound Reconstruction
Jul 05, 2024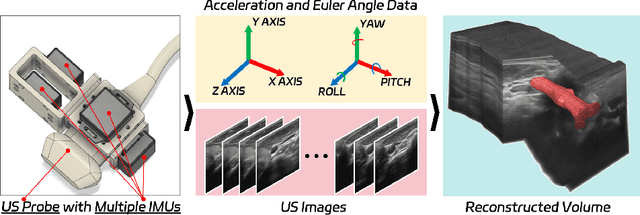


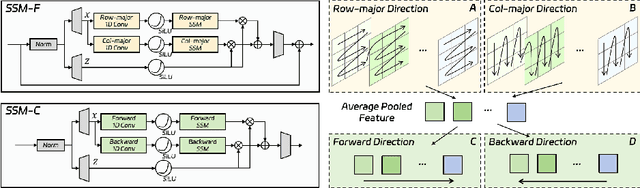
Abstract:Fine-grained spatio-temporal learning is crucial for freehand 3D ultrasound reconstruction. Previous works mainly resorted to the coarse-grained spatial features and the separated temporal dependency learning and struggles for fine-grained spatio-temporal learning. Mining spatio-temporal information in fine-grained scales is extremely challenging due to learning difficulties in long-range dependencies. In this context, we propose a novel method to exploit the long-range dependency management capabilities of the state space model (SSM) to address the above challenge. Our contribution is three-fold. First, we propose ReMamba, which mines multi-scale spatio-temporal information by devising a multi-directional SSM. Second, we propose an adaptive fusion strategy that introduces multiple inertial measurement units as auxiliary temporal information to enhance spatio-temporal perception. Last, we design an online alignment strategy that encodes the temporal information as pseudo labels for multi-modal alignment to further improve reconstruction performance. Extensive experimental validations on two large-scale datasets show remarkable improvement from our method over competitors.
FetusMapV2: Enhanced Fetal Pose Estimation in 3D Ultrasound
Oct 30, 2023Abstract:Fetal pose estimation in 3D ultrasound (US) involves identifying a set of associated fetal anatomical landmarks. Its primary objective is to provide comprehensive information about the fetus through landmark connections, thus benefiting various critical applications, such as biometric measurements, plane localization, and fetal movement monitoring. However, accurately estimating the 3D fetal pose in US volume has several challenges, including poor image quality, limited GPU memory for tackling high dimensional data, symmetrical or ambiguous anatomical structures, and considerable variations in fetal poses. In this study, we propose a novel 3D fetal pose estimation framework (called FetusMapV2) to overcome the above challenges. Our contribution is three-fold. First, we propose a heuristic scheme that explores the complementary network structure-unconstrained and activation-unreserved GPU memory management approaches, which can enlarge the input image resolution for better results under limited GPU memory. Second, we design a novel Pair Loss to mitigate confusion caused by symmetrical and similar anatomical structures. It separates the hidden classification task from the landmark localization task and thus progressively eases model learning. Last, we propose a shape priors-based self-supervised learning by selecting the relatively stable landmarks to refine the pose online. Extensive experiments and diverse applications on a large-scale fetal US dataset including 1000 volumes with 22 landmarks per volume demonstrate that our method outperforms other strong competitors.
Multi-IMU with Online Self-Consistency for Freehand 3D Ultrasound Reconstruction
Jul 19, 2023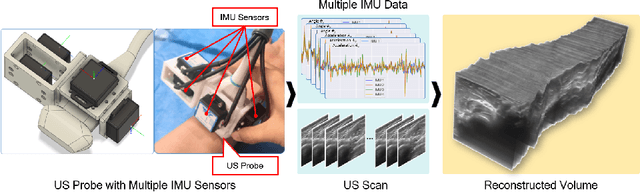
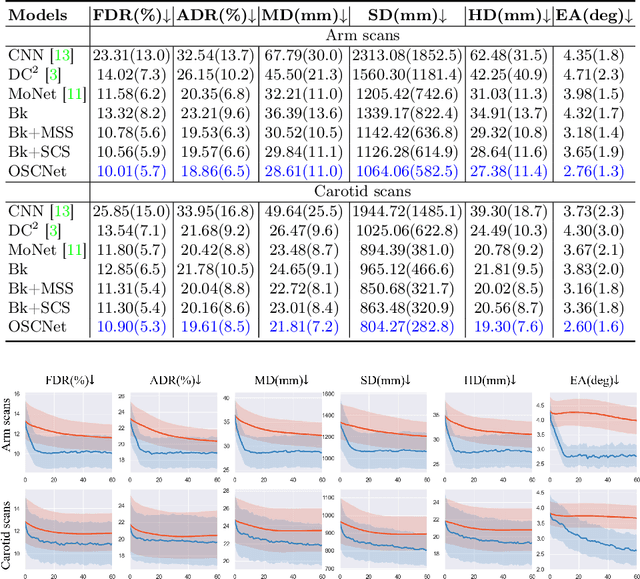
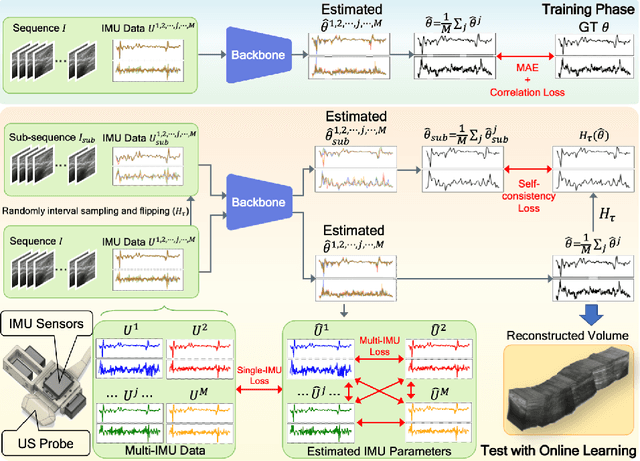
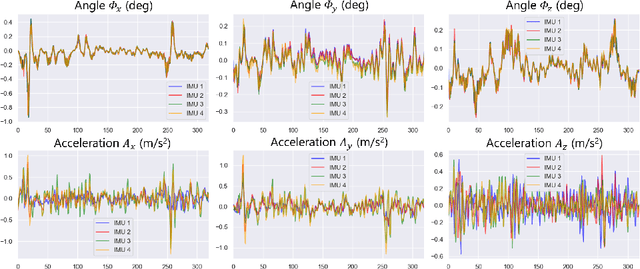
Abstract:Ultrasound (US) imaging is a popular tool in clinical diagnosis, offering safety, repeatability, and real-time capabilities. Freehand 3D US is a technique that provides a deeper understanding of scanned regions without increasing complexity. However, estimating elevation displacement and accumulation error remains challenging, making it difficult to infer the relative position using images alone. The addition of external lightweight sensors has been proposed to enhance reconstruction performance without adding complexity, which has been shown to be beneficial. We propose a novel online self-consistency network (OSCNet) using multiple inertial measurement units (IMUs) to improve reconstruction performance. OSCNet utilizes a modal-level self-supervised strategy to fuse multiple IMU information and reduce differences between reconstruction results obtained from each IMU data. Additionally, a sequence-level self-consistency strategy is proposed to improve the hierarchical consistency of prediction results among the scanning sequence and its sub-sequences. Experiments on large-scale arm and carotid datasets with multiple scanning tactics demonstrate that our OSCNet outperforms previous methods, achieving state-of-the-art reconstruction performance.
Fine-grained Correlation Loss for Regression
Jul 01, 2022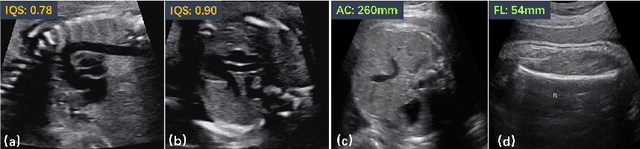
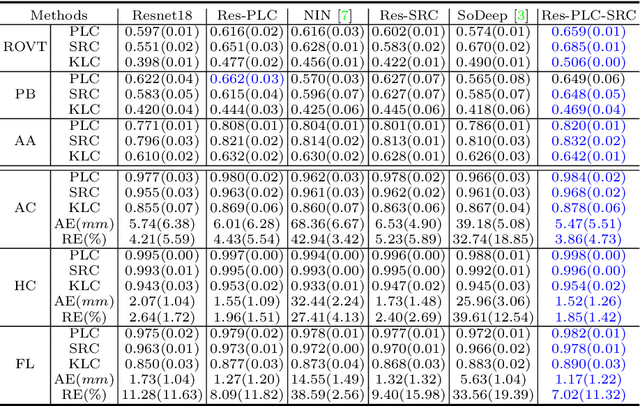
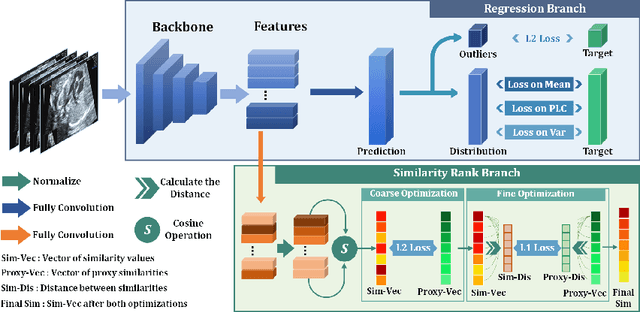
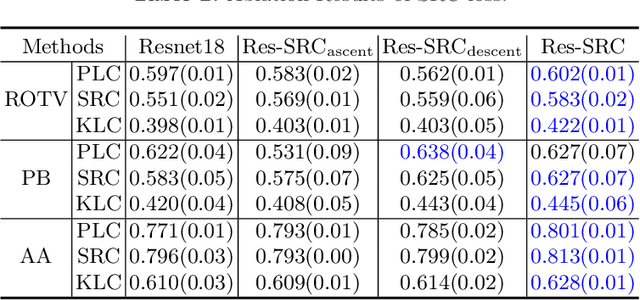
Abstract:Regression learning is classic and fundamental for medical image analysis. It provides the continuous mapping for many critical applications, like the attribute estimation, object detection, segmentation and non-rigid registration. However, previous studies mainly took the case-wise criteria, like the mean square errors, as the optimization objectives. They ignored the very important population-wise correlation criterion, which is exactly the final evaluation metric in many tasks. In this work, we propose to revisit the classic regression tasks with novel investigations on directly optimizing the fine-grained correlation losses. We mainly explore two complementary correlation indexes as learnable losses: Pearson linear correlation (PLC) and Spearman rank correlation (SRC). The contributions of this paper are two folds. First, for the PLC on global level, we propose a strategy to make it robust against the outliers and regularize the key distribution factors. These efforts significantly stabilize the learning and magnify the efficacy of PLC. Second, for the SRC on local level, we propose a coarse-to-fine scheme to ease the learning of the exact ranking order among samples. Specifically, we convert the learning for the ranking of samples into the learning of similarity relationships among samples. We extensively validate our method on two typical ultrasound image regression tasks, including the image quality assessment and bio-metric measurement. Experiments prove that, with the fine-grained guidance in directly optimizing the correlation, the regression performances are significantly improved. Our proposed correlation losses are general and can be extended to more important applications.
Deep Motion Network for Freehand 3D Ultrasound Reconstruction
Jul 01, 2022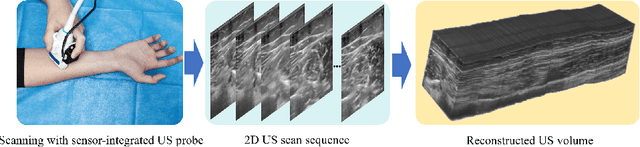
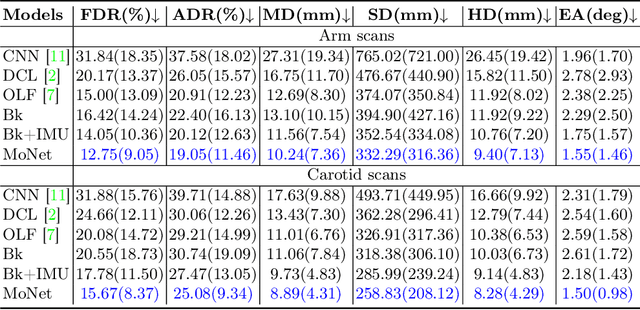
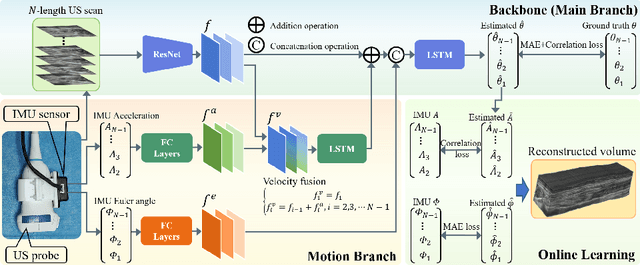
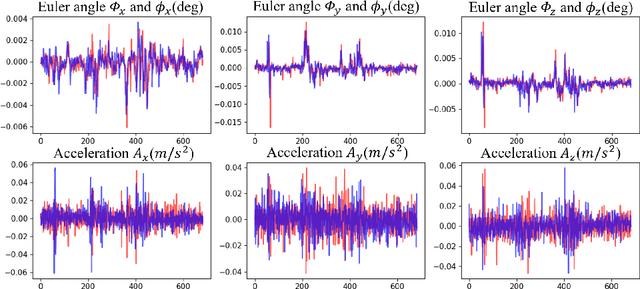
Abstract:Freehand 3D ultrasound (US) has important clinical value due to its low cost and unrestricted field of view. Recently deep learning algorithms have removed its dependence on bulky and expensive external positioning devices. However, improving reconstruction accuracy is still hampered by difficult elevational displacement estimation and large cumulative drift. In this context, we propose a novel deep motion network (MoNet) that integrates images and a lightweight sensor known as the inertial measurement unit (IMU) from a velocity perspective to alleviate the obstacles mentioned above. Our contribution is two-fold. First, we introduce IMU acceleration for the first time to estimate elevational displacements outside the plane. We propose a temporal and multi-branch structure to mine the valuable information of low signal-to-noise ratio (SNR) acceleration. Second, we propose a multi-modal online self-supervised strategy that leverages IMU information as weak labels for adaptive optimization to reduce drift errors and further ameliorate the impacts of acceleration noise. Experiments show that our proposed method achieves the superior reconstruction performance, exceeding state-of-the-art methods across the board.
HASA: Hybrid Architecture Search with Aggregation Strategy for Echinococcosis Classification and Ovary Segmentation in Ultrasound Images
Apr 20, 2022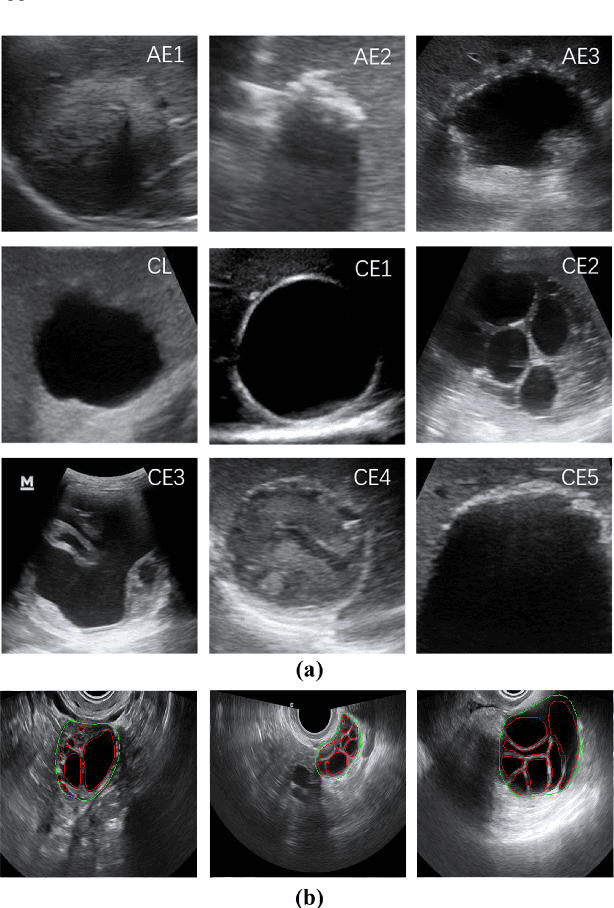
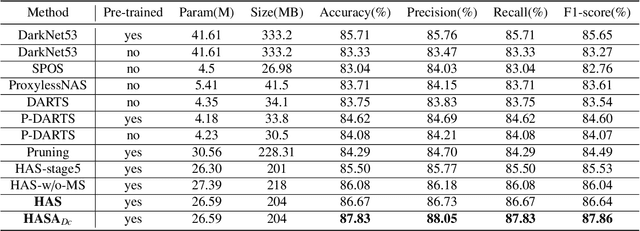
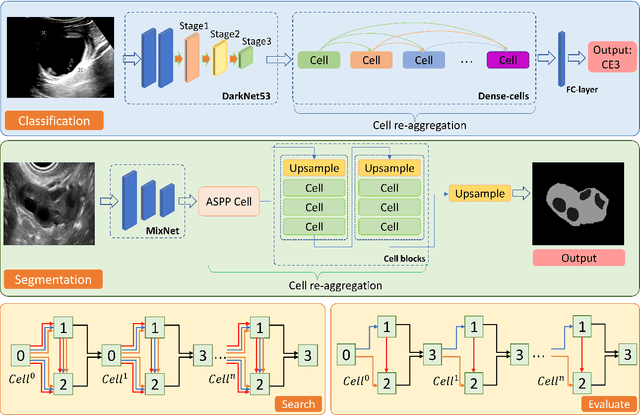
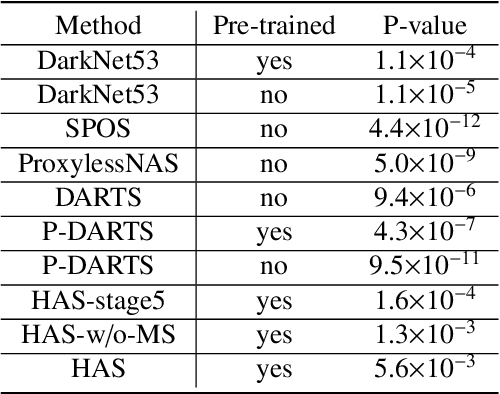
Abstract:Different from handcrafted features, deep neural networks can automatically learn task-specific features from data. Due to this data-driven nature, they have achieved remarkable success in various areas. However, manual design and selection of suitable network architectures are time-consuming and require substantial effort of human experts. To address this problem, researchers have proposed neural architecture search (NAS) algorithms which can automatically generate network architectures but suffer from heavy computational cost and instability if searching from scratch. In this paper, we propose a hybrid NAS framework for ultrasound (US) image classification and segmentation. The hybrid framework consists of a pre-trained backbone and several searched cells (i.e., network building blocks), which takes advantage of the strengths of both NAS and the expert knowledge from existing convolutional neural networks. Specifically, two effective and lightweight operations, a mixed depth-wise convolution operator and a squeeze-and-excitation block, are introduced into the candidate operations to enhance the variety and capacity of the searched cells. These two operations not only decrease model parameters but also boost network performance. Moreover, we propose a re-aggregation strategy for the searched cells, aiming to further improve the performance for different vision tasks. We tested our method on two large US image datasets, including a 9-class echinococcosis dataset containing 9566 images for classification and an ovary dataset containing 3204 images for segmentation. Ablation experiments and comparison with other handcrafted or automatically searched architectures demonstrate that our method can generate more powerful and lightweight models for the above US image classification and segmentation tasks.
Self Context and Shape Prior for Sensorless Freehand 3D Ultrasound Reconstruction
Aug 18, 2021



Abstract:3D ultrasound (US) is widely used for its rich diagnostic information. However, it is criticized for its limited field of view. 3D freehand US reconstruction is promising in addressing the problem by providing broad range and freeform scan. The existing deep learning based methods only focus on the basic cases of skill sequences, and the model relies on the training data heavily. The sequences in real clinical practice are a mix of diverse skills and have complex scanning paths. Besides, deep models should adapt themselves to the testing cases with prior knowledge for better robustness, rather than only fit to the training cases. In this paper, we propose a novel approach to sensorless freehand 3D US reconstruction considering the complex skill sequences. Our contribution is three-fold. First, we advance a novel online learning framework by designing a differentiable reconstruction algorithm. It realizes an end-to-end optimization from section sequences to the reconstructed volume. Second, a self-supervised learning method is developed to explore the context information that reconstructed by the testing data itself, promoting the perception of the model. Third, inspired by the effectiveness of shape prior, we also introduce adversarial training to strengthen the learning of anatomical shape prior in the reconstructed volume. By mining the context and structural cues of the testing data, our online learning methods can drive the model to handle complex skill sequences. Experimental results on developmental dysplasia of the hip US and fetal US datasets show that, our proposed method can outperform the start-of-the-art methods regarding the shift errors and path similarities.
Style-invariant Cardiac Image Segmentation with Test-time Augmentation
Sep 24, 2020



Abstract:Deep models often suffer from severe performance drop due to the appearance shift in the real clinical setting. Most of the existing learning-based methods rely on images from multiple sites/vendors or even corresponding labels. However, collecting enough unknown data to robustly model segmentation cannot always hold since the complex appearance shift caused by imaging factors in daily application. In this paper, we propose a novel style-invariant method for cardiac image segmentation. Based on the zero-shot style transfer to remove appearance shift and test-time augmentation to explore diverse underlying anatomy, our proposed method is effective in combating the appearance shift. Our contribution is three-fold. First, inspired by the spirit of universal style transfer, we develop a zero-shot stylization for content images to generate stylized images that appearance similarity to the style images. Second, we build up a robust cardiac segmentation model based on the U-Net structure. Our framework mainly consists of two networks during testing: the ST network for removing appearance shift and the segmentation network. Third, we investigate test-time augmentation to explore transformed versions of the stylized image for prediction and the results are merged. Notably, our proposed framework is fully test-time adaptation. Experiment results demonstrate that our methods are promising and generic for generalizing deep segmentation models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge