Tong Han
Mitral Regurgitation Recogniton based on Unsupervised Out-of-Distribution Detection with Residual Diffusion Amplification
Jul 31, 2024



Abstract:Mitral regurgitation (MR) is a serious heart valve disease. Early and accurate diagnosis of MR via ultrasound video is critical for timely clinical decision-making and surgical intervention. However, manual MR diagnosis heavily relies on the operator's experience, which may cause misdiagnosis and inter-observer variability. Since MR data is limited and has large intra-class variability, we propose an unsupervised out-of-distribution (OOD) detection method to identify MR rather than building a deep classifier. To our knowledge, we are the first to explore OOD in MR ultrasound videos. Our method consists of a feature extractor, a feature reconstruction model, and a residual accumulation amplification algorithm. The feature extractor obtains features from the video clips and feeds them into the feature reconstruction model to restore the original features. The residual accumulation amplification algorithm then iteratively performs noise feature reconstruction, amplifying the reconstructed error of OOD features. This algorithm is straightforward yet efficient and can seamlessly integrate as a plug-and-play component in reconstruction-based OOD detection methods. We validated the proposed method on a large ultrasound dataset containing 893 non-MR and 267 MR videos. Experimental results show that our OOD detection method can effectively identify MR samples.
Completed Feature Disentanglement Learning for Multimodal MRIs Analysis
Jul 06, 2024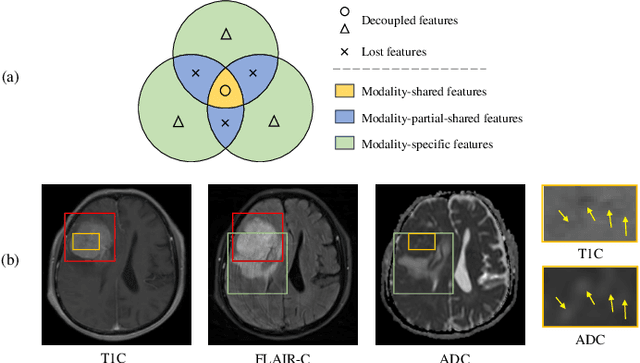
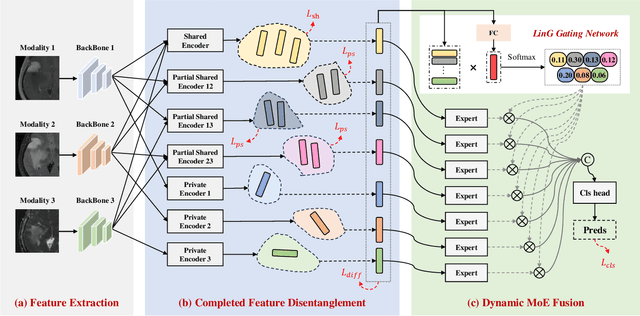
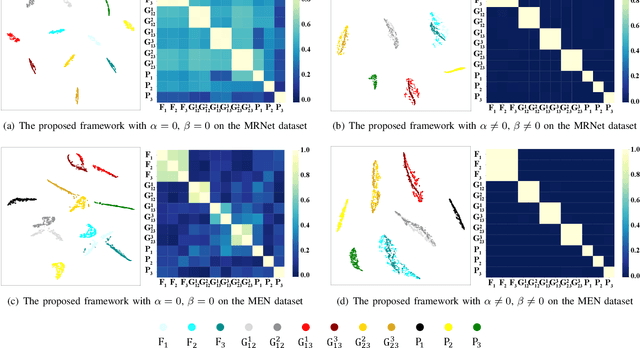
Abstract:Multimodal MRIs play a crucial role in clinical diagnosis and treatment. Feature disentanglement (FD)-based methods, aiming at learning superior feature representations for multimodal data analysis, have achieved significant success in multimodal learning (MML). Typically, existing FD-based methods separate multimodal data into modality-shared and modality-specific features, and employ concatenation or attention mechanisms to integrate these features. However, our preliminary experiments indicate that these methods could lead to a loss of shared information among subsets of modalities when the inputs contain more than two modalities, and such information is critical for prediction accuracy. Furthermore, these methods do not adequately interpret the relationships between the decoupled features at the fusion stage. To address these limitations, we propose a novel Complete Feature Disentanglement (CFD) strategy that recovers the lost information during feature decoupling. Specifically, the CFD strategy not only identifies modality-shared and modality-specific features, but also decouples shared features among subsets of multimodal inputs, termed as modality-partial-shared features. We further introduce a new Dynamic Mixture-of-Experts Fusion (DMF) module that dynamically integrates these decoupled features, by explicitly learning the local-global relationships among the features. The effectiveness of our approach is validated through classification tasks on three multimodal MRI datasets. Extensive experimental results demonstrate that our approach outperforms other state-of-the-art MML methods with obvious margins, showcasing its superior performance.
A Foundation Model for General Moving Object Segmentation in Medical Images
Oct 04, 2023



Abstract:Medical image segmentation aims to delineate the anatomical or pathological structures of interest, playing a crucial role in clinical diagnosis. A substantial amount of high-quality annotated data is crucial for constructing high-precision deep segmentation models. However, medical annotation is highly cumbersome and time-consuming, especially for medical videos or 3D volumes, due to the huge labeling space and poor inter-frame consistency. Recently, a fundamental task named Moving Object Segmentation (MOS) has made significant advancements in natural images. Its objective is to delineate moving objects from the background within image sequences, requiring only minimal annotations. In this paper, we propose the first foundation model, named iMOS, for MOS in medical images. Extensive experiments on a large multi-modal medical dataset validate the effectiveness of the proposed iMOS. Specifically, with the annotation of only a small number of images in the sequence, iMOS can achieve satisfactory tracking and segmentation performance of moving objects throughout the entire sequence in bi-directions. We hope that the proposed iMOS can help accelerate the annotation speed of experts, and boost the development of medical foundation models.
OnUVS: Online Feature Decoupling Framework for High-Fidelity Ultrasound Video Synthesis
Aug 16, 2023



Abstract:Ultrasound (US) imaging is indispensable in clinical practice. To diagnose certain diseases, sonographers must observe corresponding dynamic anatomic structures to gather comprehensive information. However, the limited availability of specific US video cases causes teaching difficulties in identifying corresponding diseases, which potentially impacts the detection rate of such cases. The synthesis of US videos may represent a promising solution to this issue. Nevertheless, it is challenging to accurately animate the intricate motion of dynamic anatomic structures while preserving image fidelity. To address this, we present a novel online feature-decoupling framework called OnUVS for high-fidelity US video synthesis. Our highlights can be summarized by four aspects. First, we introduced anatomic information into keypoint learning through a weakly-supervised training strategy, resulting in improved preservation of anatomical integrity and motion while minimizing the labeling burden. Second, to better preserve the integrity and textural information of US images, we implemented a dual-decoder that decouples the content and textural features in the generator. Third, we adopted a multiple-feature discriminator to extract a comprehensive range of visual cues, thereby enhancing the sharpness and fine details of the generated videos. Fourth, we constrained the motion trajectories of keypoints during online learning to enhance the fluidity of generated videos. Our validation and user studies on in-house echocardiographic and pelvic floor US videos showed that OnUVS synthesizes US videos with high fidelity.
Joint Prediction of Meningioma Grade and Brain Invasion via Task-Aware Contrastive Learning
Sep 04, 2022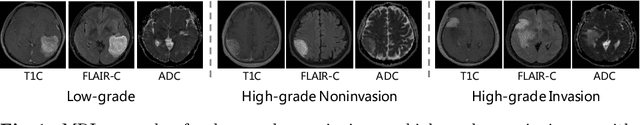
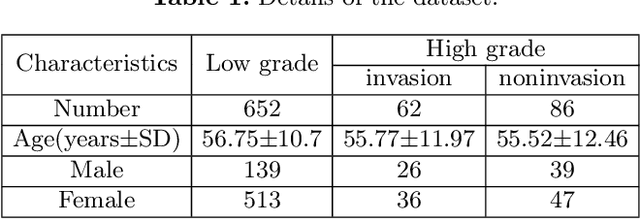
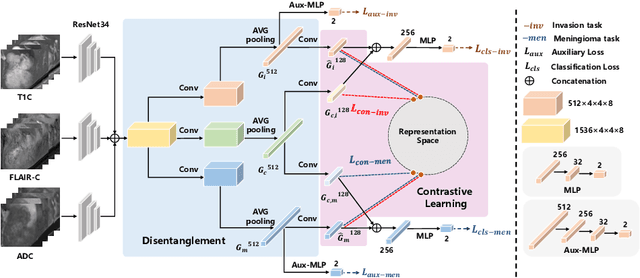
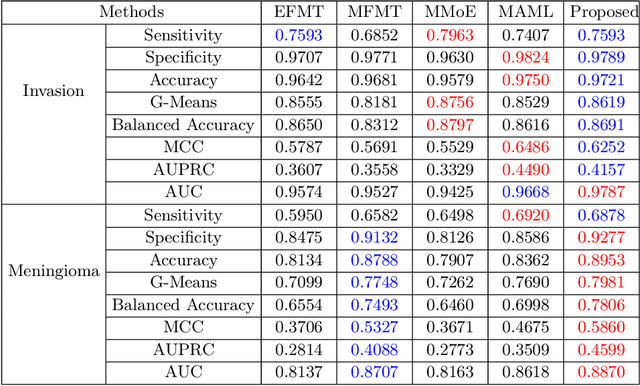
Abstract:Preoperative and noninvasive prediction of the meningioma grade is important in clinical practice, as it directly influences the clinical decision making. What's more, brain invasion in meningioma (i.e., the presence of tumor tissue within the adjacent brain tissue) is an independent criterion for the grading of meningioma and influences the treatment strategy. Although efforts have been reported to address these two tasks, most of them rely on hand-crafted features and there is no attempt to exploit the two prediction tasks simultaneously. In this paper, we propose a novel task-aware contrastive learning algorithm to jointly predict meningioma grade and brain invasion from multi-modal MRIs. Based on the basic multi-task learning framework, our key idea is to adopt contrastive learning strategy to disentangle the image features into task-specific features and task-common features, and explicitly leverage their inherent connections to improve feature representation for the two prediction tasks. In this retrospective study, an MRI dataset was collected, for which 800 patients (containing 148 high-grade, 62 invasion) were diagnosed with meningioma by pathological analysis. Experimental results show that the proposed algorithm outperforms alternative multi-task learning methods, achieving AUCs of 0:8870 and 0:9787 for the prediction of meningioma grade and brain invasion, respectively. The code is available at https://github.com/IsDling/predictTCL.
NestedFormer: Nested Modality-Aware Transformer for Brain Tumor Segmentation
Aug 31, 2022



Abstract:Multi-modal MR imaging is routinely used in clinical practice to diagnose and investigate brain tumors by providing rich complementary information. Previous multi-modal MRI segmentation methods usually perform modal fusion by concatenating multi-modal MRIs at an early/middle stage of the network, which hardly explores non-linear dependencies between modalities. In this work, we propose a novel Nested Modality-Aware Transformer (NestedFormer) to explicitly explore the intra-modality and inter-modality relationships of multi-modal MRIs for brain tumor segmentation. Built on the transformer-based multi-encoder and single-decoder structure, we perform nested multi-modal fusion for high-level representations of different modalities and apply modality-sensitive gating (MSG) at lower scales for more effective skip connections. Specifically, the multi-modal fusion is conducted in our proposed Nested Modality-aware Feature Aggregation (NMaFA) module, which enhances long-term dependencies within individual modalities via a tri-orientated spatial-attention transformer, and further complements key contextual information among modalities via a cross-modality attention transformer. Extensive experiments on BraTS2020 benchmark and a private meningiomas segmentation (MeniSeg) dataset show that the NestedFormer clearly outperforms the state-of-the-arts. The code is available at https://github.com/920232796/NestedFormer.
Towards Clinical Diagnosis: Automated Stroke Lesion Segmentation on Multimodal MR Image Using Convolutional Neural Network
Mar 05, 2018



Abstract:The patient with ischemic stroke can benefit most from the earliest possible definitive diagnosis. While the high quality medical resources are quite scarce across the globe, an automated diagnostic tool is expected in analyzing the magnetic resonance (MR) images to provide reference in clinical diagnosis. In this paper, we propose a deep learning method to automatically segment ischemic stroke lesions from multi-modal MR images. By using atrous convolution and global convolution network, our proposed residual-structured fully convolutional network (Res-FCN) is able to capture features from large receptive fields. The network architecture is validated on a large dataset of 212 clinically acquired multi-modal MR images, which is shown to achieve a mean dice coefficient of 0.645 with a mean number of false negative lesions of 1.515. The false negatives can reach a value that close to a common medical image doctor, making it exceptive for a real clinical application.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge