Shuxue Ding
SCANet: Split Coordinate Attention Network for Building Footprint Extraction
Jul 28, 2025Abstract:Building footprint extraction holds immense significance in remote sensing image analysis and has great value in urban planning, land use, environmental protection and disaster assessment. Despite the progress made by conventional and deep learning approaches in this field, they continue to encounter significant challenges. This paper introduces a novel plug-and-play attention module, Split Coordinate Attention (SCA), which ingeniously captures spatially remote interactions by employing two spatial range of pooling kernels, strategically encoding each channel along x and y planes, and separately performs a series of split operations for each feature group, thus enabling more efficient semantic feature extraction. By inserting into a 2D CNN to form an effective SCANet, our SCANet outperforms recent SOTA methods on the public Wuhan University (WHU) Building Dataset and Massachusetts Building Dataset in terms of various metrics. Particularly SCANet achieves the best IoU, 91.61% and 75.49% for the two datasets. Our code is available at https://github.com/AiEson/SCANet
A Multi-Stage Framework for 3D Individual Tooth Segmentation in Dental CBCT
Jul 15, 2024Abstract:Cone beam computed tomography (CBCT) is a common way of diagnosing dental related diseases. Accurate segmentation of 3D tooth is of importance for the treatment. Although deep learning based methods have achieved convincing results in medical image processing, they need a large of annotated data for network training, making it very time-consuming in data collection and annotation. Besides, domain shift widely existing in the distribution of data acquired by different devices impacts severely the model generalization. To resolve the problem, we propose a multi-stage framework for 3D tooth segmentation in dental CBCT, which achieves the third place in the "Semi-supervised Teeth Segmentation" 3D (STS-3D) challenge. The experiments on validation set compared with other semi-supervised segmentation methods further indicate the validity of our approach.
CrossMatch: Enhance Semi-Supervised Medical Image Segmentation with Perturbation Strategies and Knowledge Distillation
May 01, 2024Abstract:Semi-supervised learning for medical image segmentation presents a unique challenge of efficiently using limited labeled data while leveraging abundant unlabeled data. Despite advancements, existing methods often do not fully exploit the potential of the unlabeled data for enhancing model robustness and accuracy. In this paper, we introduce CrossMatch, a novel framework that integrates knowledge distillation with dual perturbation strategies-image-level and feature-level-to improve the model's learning from both labeled and unlabeled data. CrossMatch employs multiple encoders and decoders to generate diverse data streams, which undergo self-knowledge distillation to enhance consistency and reliability of predictions across varied perturbations. Our method significantly surpasses other state-of-the-art techniques in standard benchmarks by effectively minimizing the gap between training on labeled and unlabeled data and improving edge accuracy and generalization in medical image segmentation. The efficacy of CrossMatch is demonstrated through extensive experimental validations, showing remarkable performance improvements without increasing computational costs. Code for this implementation is made available at https://github.com/AiEson/CrossMatch.git.
Automatic acute ischemic stroke lesion segmentation using semi-supervised learning
Aug 10, 2019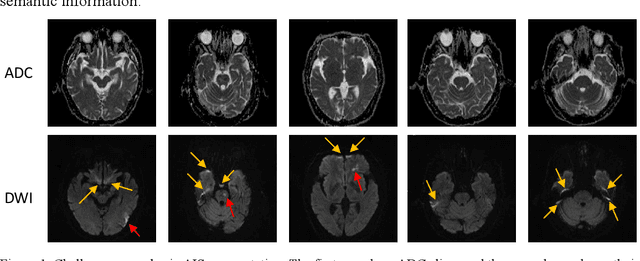


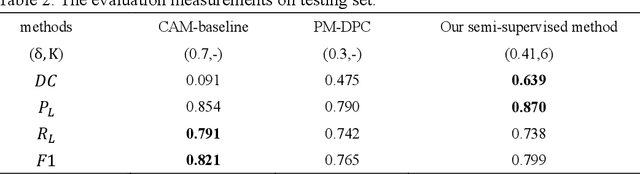
Abstract:Ischemic stroke is a common disease in the elderly population, which can cause long-term disability and even death. However, the time window for treatment of ischemic stroke in its acute stage is very short. To fast localize and quantitively evaluate the acute ischemic stroke (AIS) lesions, many deep-learning-based lesion segmentation methods have been proposed in the literature, where a deep convolutional neural network (CNN) was trained on hundreds of fully labeled subjects with accurate annotations of AIS lesions. Despite that high segmentation accuracy can be achieved, the accurate labels should be annotated by experienced clinicians, and it is therefore very time-consuming to obtain a large number of fully labeled subjects. In this paper, we propose a semi-supervised method to automatically segment AIS lesions in diffusion weighted images and apparent diffusion coefficient maps. By using a large number of weakly labeled subjects and a small number of fully labeled subjects, our proposed method is able to accurately detect and segment the AIS lesions. In particular, our proposed method consists of three parts: 1) a double-path classification net (DPC-Net) trained in a weakly-supervised way is used to detect the suspicious regions of AIS lesions; 2) a pixel-level K-Means clustering algorithm is used to identify the hyperintensive regions on the DWIs; and 3) a region-growing algorithm combines the outputs of the DPC-Net and the K-Means to obtain the final precise lesion segmentation. In our experiment, we use 460 weakly labeled subjects and 15 fully labeled subjects to train and fine-tune the proposed method. By evaluating on a clinical dataset with 150 fully labeled subjects, our proposed method achieves a mean dice coefficient of 0.639, and a lesion-wise F1 score of 0.799.
Towards Clinical Diagnosis: Automated Stroke Lesion Segmentation on Multimodal MR Image Using Convolutional Neural Network
Mar 05, 2018



Abstract:The patient with ischemic stroke can benefit most from the earliest possible definitive diagnosis. While the high quality medical resources are quite scarce across the globe, an automated diagnostic tool is expected in analyzing the magnetic resonance (MR) images to provide reference in clinical diagnosis. In this paper, we propose a deep learning method to automatically segment ischemic stroke lesions from multi-modal MR images. By using atrous convolution and global convolution network, our proposed residual-structured fully convolutional network (Res-FCN) is able to capture features from large receptive fields. The network architecture is validated on a large dataset of 212 clinically acquired multi-modal MR images, which is shown to achieve a mean dice coefficient of 0.645 with a mean number of false negative lesions of 1.515. The false negatives can reach a value that close to a common medical image doctor, making it exceptive for a real clinical application.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge