Tianling Liu
Completed Feature Disentanglement Learning for Multimodal MRIs Analysis
Jul 06, 2024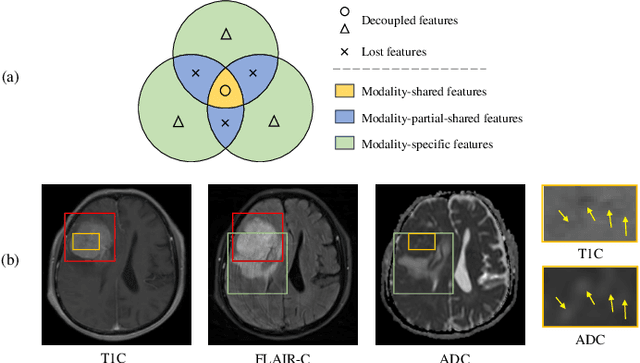
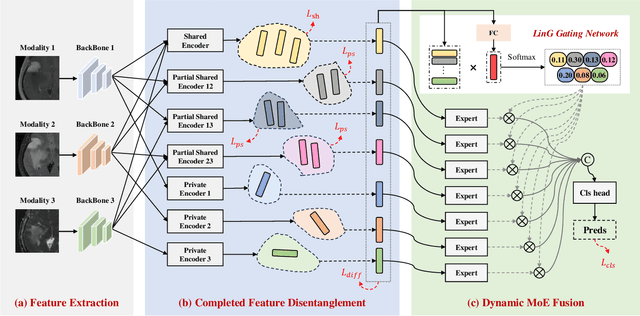
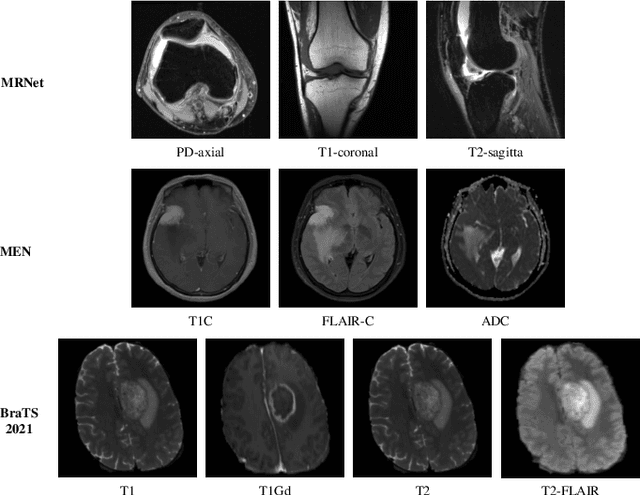
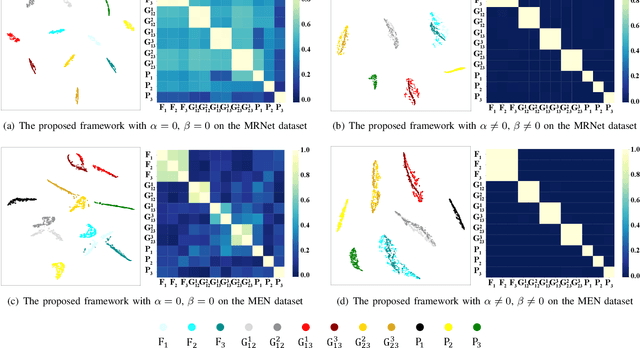
Abstract:Multimodal MRIs play a crucial role in clinical diagnosis and treatment. Feature disentanglement (FD)-based methods, aiming at learning superior feature representations for multimodal data analysis, have achieved significant success in multimodal learning (MML). Typically, existing FD-based methods separate multimodal data into modality-shared and modality-specific features, and employ concatenation or attention mechanisms to integrate these features. However, our preliminary experiments indicate that these methods could lead to a loss of shared information among subsets of modalities when the inputs contain more than two modalities, and such information is critical for prediction accuracy. Furthermore, these methods do not adequately interpret the relationships between the decoupled features at the fusion stage. To address these limitations, we propose a novel Complete Feature Disentanglement (CFD) strategy that recovers the lost information during feature decoupling. Specifically, the CFD strategy not only identifies modality-shared and modality-specific features, but also decouples shared features among subsets of multimodal inputs, termed as modality-partial-shared features. We further introduce a new Dynamic Mixture-of-Experts Fusion (DMF) module that dynamically integrates these decoupled features, by explicitly learning the local-global relationships among the features. The effectiveness of our approach is validated through classification tasks on three multimodal MRI datasets. Extensive experimental results demonstrate that our approach outperforms other state-of-the-art MML methods with obvious margins, showcasing its superior performance.
EOCSA: Predicting Prognosis of Epithelial Ovarian Cancer with Whole Slide Histopathological Images
Oct 11, 2022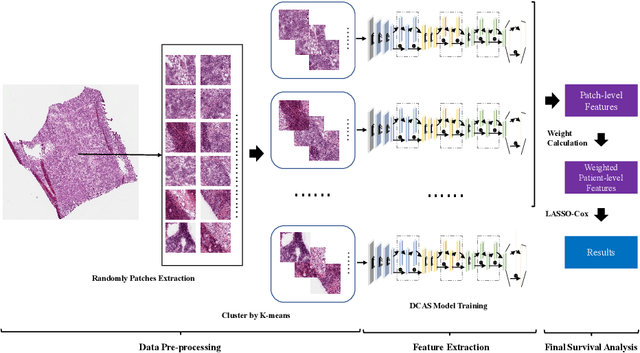
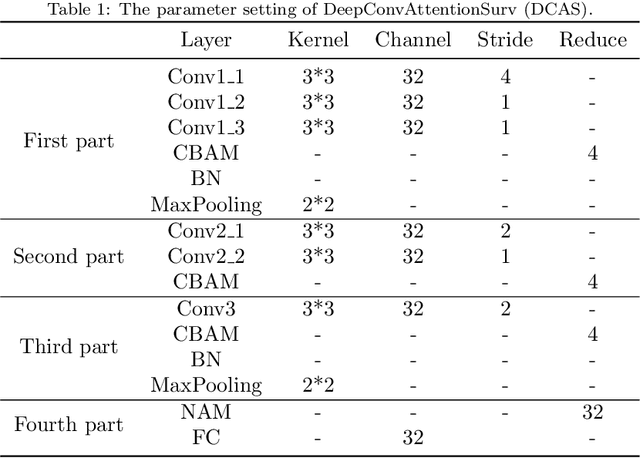
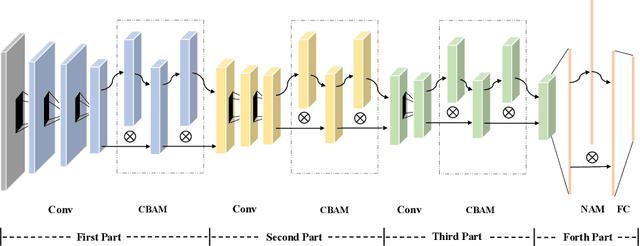

Abstract:Ovarian cancer is one of the most serious cancers that threaten women around the world. Epithelial ovarian cancer (EOC), as the most commonly seen subtype of ovarian cancer, has rather high mortality rate and poor prognosis among various gynecological cancers. Survival analysis outcome is able to provide treatment advices to doctors. In recent years, with the development of medical imaging technology, survival prediction approaches based on pathological images have been proposed. In this study, we designed a deep framework named EOCSA which analyzes the prognosis of EOC patients based on pathological whole slide images (WSIs). Specifically, we first randomly extracted patches from WSIs and grouped them into multiple clusters. Next, we developed a survival prediction model, named DeepConvAttentionSurv (DCAS), which was able to extract patch-level features, removed less discriminative clusters and predicted the EOC survival precisely. Particularly, channel attention, spatial attention, and neuron attention mechanisms were used to improve the performance of feature extraction. Then patient-level features were generated from our weight calculation method and the survival time was finally estimated using LASSO-Cox model. The proposed EOCSA is efficient and effective in predicting prognosis of EOC and the DCAS ensures more informative and discriminative features can be extracted. As far as we know, our work is the first to analyze the survival of EOC based on WSIs and deep neural network technologies. The experimental results demonstrate that our proposed framework has achieved state-of-the-art performance of 0.980 C-index. The implementation of the approach can be found at https://github.com/RanSuLab/EOCprognosis.
Joint Prediction of Meningioma Grade and Brain Invasion via Task-Aware Contrastive Learning
Sep 04, 2022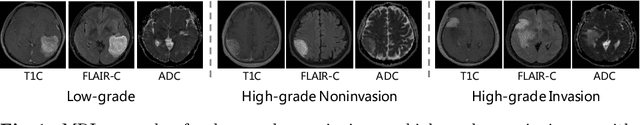
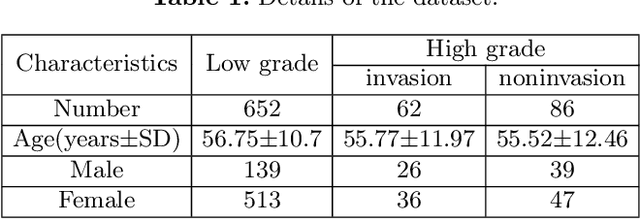
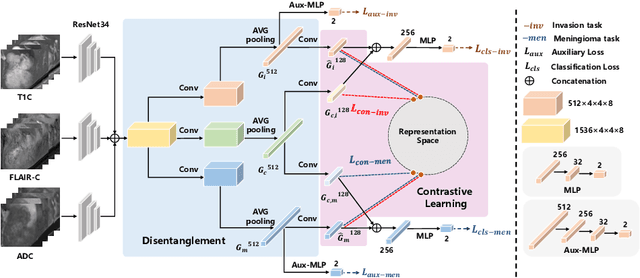
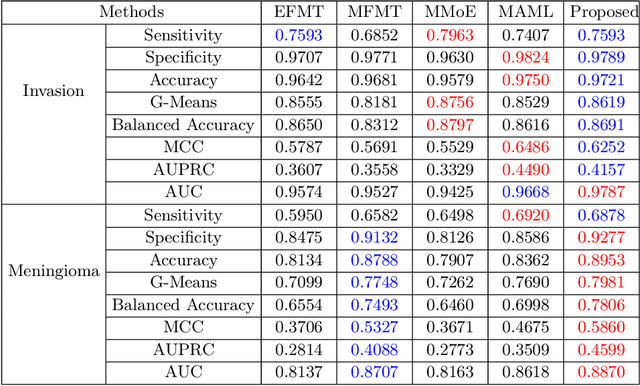
Abstract:Preoperative and noninvasive prediction of the meningioma grade is important in clinical practice, as it directly influences the clinical decision making. What's more, brain invasion in meningioma (i.e., the presence of tumor tissue within the adjacent brain tissue) is an independent criterion for the grading of meningioma and influences the treatment strategy. Although efforts have been reported to address these two tasks, most of them rely on hand-crafted features and there is no attempt to exploit the two prediction tasks simultaneously. In this paper, we propose a novel task-aware contrastive learning algorithm to jointly predict meningioma grade and brain invasion from multi-modal MRIs. Based on the basic multi-task learning framework, our key idea is to adopt contrastive learning strategy to disentangle the image features into task-specific features and task-common features, and explicitly leverage their inherent connections to improve feature representation for the two prediction tasks. In this retrospective study, an MRI dataset was collected, for which 800 patients (containing 148 high-grade, 62 invasion) were diagnosed with meningioma by pathological analysis. Experimental results show that the proposed algorithm outperforms alternative multi-task learning methods, achieving AUCs of 0:8870 and 0:9787 for the prediction of meningioma grade and brain invasion, respectively. The code is available at https://github.com/IsDling/predictTCL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge