Shengli Li
Simple is what you need for efficient and accurate medical image segmentation
Jun 16, 2025Abstract:While modern segmentation models often prioritize performance over practicality, we advocate a design philosophy prioritizing simplicity and efficiency, and attempted high performance segmentation model design. This paper presents SimpleUNet, a scalable ultra-lightweight medical image segmentation model with three key innovations: (1) A partial feature selection mechanism in skip connections for redundancy reduction while enhancing segmentation performance; (2) A fixed-width architecture that prevents exponential parameter growth across network stages; (3) An adaptive feature fusion module achieving enhanced representation with minimal computational overhead. With a record-breaking 16 KB parameter configuration, SimpleUNet outperforms LBUNet and other lightweight benchmarks across multiple public datasets. The 0.67 MB variant achieves superior efficiency (8.60 GFLOPs) and accuracy, attaining a mean DSC/IoU of 85.76%/75.60% on multi-center breast lesion datasets, surpassing both U-Net and TransUNet. Evaluations on skin lesion datasets (ISIC 2017/2018: mDice 84.86%/88.77%) and endoscopic polyp segmentation (KVASIR-SEG: 86.46%/76.48% mDice/mIoU) confirm consistent dominance over state-of-the-art models. This work demonstrates that extreme model compression need not compromise performance, providing new insights for efficient and accurate medical image segmentation. Codes can be found at https://github.com/Frankyu5666666/SimpleUNet.
FPDANet: A Multi-Section Classification Model for Intelligent Screening of Fetal Ultrasound
Jun 06, 2025Abstract:ResNet has been widely used in image classification tasks due to its ability to model the residual dependence of constant mappings for linear computation. However, the ResNet method adopts a unidirectional transfer of features and lacks an effective method to correlate contextual information, which is not effective in classifying fetal ultrasound images in the classification task, and fetal ultrasound images have problems such as low contrast, high similarity, and high noise. Therefore, we propose a bilateral multi-scale information fusion network-based FPDANet to address the above challenges. Specifically, we design the positional attention mechanism (DAN) module, which utilizes the similarity of features to establish the dependency of different spatial positional features and enhance the feature representation. In addition, we design a bilateral multi-scale (FPAN) information fusion module to capture contextual and global feature dependencies at different feature scales, thereby further improving the model representation. FPDANet classification results obtained 91.05\% and 100\% in Top-1 and Top-5 metrics, respectively, and the experimental results proved the effectiveness and robustness of FPDANet.
Hybrid Attention for Automatic Segmentation of Whole Fetal Head in Prenatal Ultrasound Volumes
Apr 28, 2020



Abstract:Background and Objective: Biometric measurements of fetal head are important indicators for maternal and fetal health monitoring during pregnancy. 3D ultrasound (US) has unique advantages over 2D scan in covering the whole fetal head and may promote the diagnoses. However, automatically segmenting the whole fetal head in US volumes still pends as an emerging and unsolved problem. The challenges that automated solutions need to tackle include the poor image quality, boundary ambiguity, long-span occlusion, and the appearance variability across different fetal poses and gestational ages. In this paper, we propose the first fully-automated solution to segment the whole fetal head in US volumes. Methods: The segmentation task is firstly formulated as an end-to-end volumetric mapping under an encoder-decoder deep architecture. We then combine the segmentor with a proposed hybrid attention scheme (HAS) to select discriminative features and suppress the non-informative volumetric features in a composite and hierarchical way. With little computation overhead, HAS proves to be effective in addressing boundary ambiguity and deficiency. To enhance the spatial consistency in segmentation, we further organize multiple segmentors in a cascaded fashion to refine the results by revisiting context in the prediction of predecessors. Results: Validated on a large dataset collected from 100 healthy volunteers, our method presents superior segmentation performance (DSC (Dice Similarity Coefficient), 96.05%), remarkable agreements with experts. With another 156 volumes collected from 52 volunteers, we ahieve high reproducibilities (mean standard deviation 11.524 mL) against scan variations. Conclusion: This is the first investigation about whole fetal head segmentation in 3D US. Our method is promising to be a feasible solution in assisting the volumetric US-based prenatal studies.
FetusMap: Fetal Pose Estimation in 3D Ultrasound
Oct 11, 2019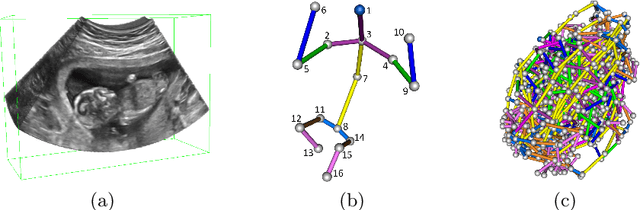
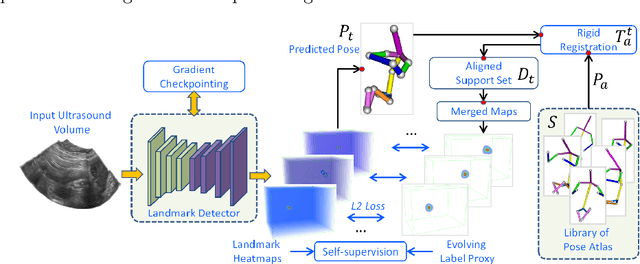
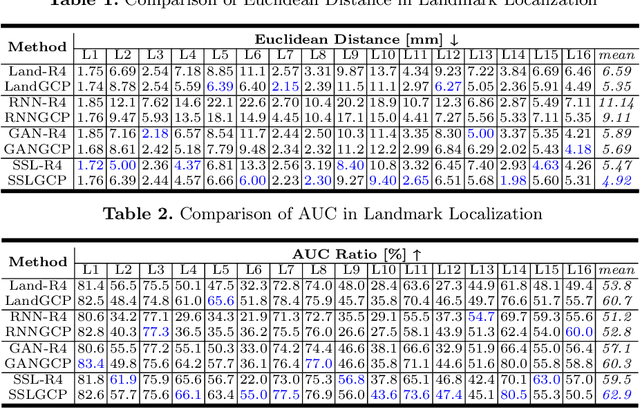
Abstract:The 3D ultrasound (US) entrance inspires a multitude of automated prenatal examinations. However, studies about the structuralized description of the whole fetus in 3D US are still rare. In this paper, we propose to estimate the 3D pose of fetus in US volumes to facilitate its quantitative analyses in global and local scales. Given the great challenges in 3D US, including the high volume dimension, poor image quality, symmetric ambiguity in anatomical structures and large variations of fetal pose, our contribution is three-fold. (i) This is the first work about 3D pose estimation of fetus in the literature. We aim to extract the skeleton of whole fetus and assign different segments/joints with correct torso/limb labels. (ii) We propose a self-supervised learning (SSL) framework to finetune the deep network to form visually plausible pose predictions. Specifically, we leverage the landmark-based registration to effectively encode case-adaptive anatomical priors and generate evolving label proxy for supervision. (iii) To enable our 3D network perceive better contextual cues with higher resolution input under limited computing resource, we further adopt the gradient check-pointing (GCP) strategy to save GPU memory and improve the prediction. Extensively validated on a large 3D US dataset, our method tackles varying fetal poses and achieves promising results. 3D pose estimation of fetus has potentials in serving as a map to provide navigation for many advanced studies.
Joint Segmentation and Landmark Localization of Fetal Femur in Ultrasound Volumes
Aug 31, 2019



Abstract:Volumetric ultrasound has great potentials in promoting prenatal examinations. Automated solutions are highly desired to efficiently and effectively analyze the massive volumes. Segmentation and landmark localization are two key techniques in making the quantitative evaluation of prenatal ultrasound volumes available in clinic. However, both tasks are non-trivial when considering the poor image quality, boundary ambiguity and anatomical variations in volumetric ultrasound. In this paper, we propose an effective framework for simultaneous segmentation and landmark localization in prenatal ultrasound volumes. The proposed framework has two branches where informative cues of segmentation and landmark localization can be propagated bidirectionally to benefit both tasks. As landmark localization tends to suffer from false positives, we propose a distance based loss to suppress the noise and thus enhance the localization map and in turn the segmentation. Finally, we further leverage an adversarial module to emphasize the correspondence between segmentation and landmark localization. Extensively validated on a volumetric ultrasound dataset of fetal femur, our proposed framework proves to be a promising solution to facilitate the interpretation of prenatal ultrasound volumes.
Deep Adaptive Network: An Efficient Deep Neural Network with Sparse Binary Connections
Apr 21, 2016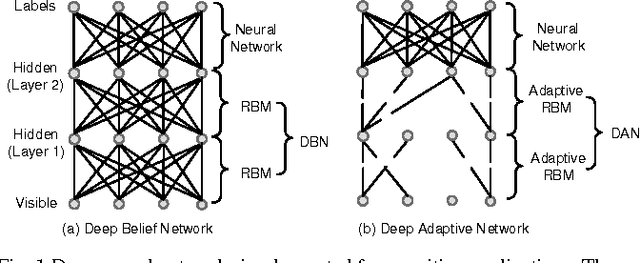
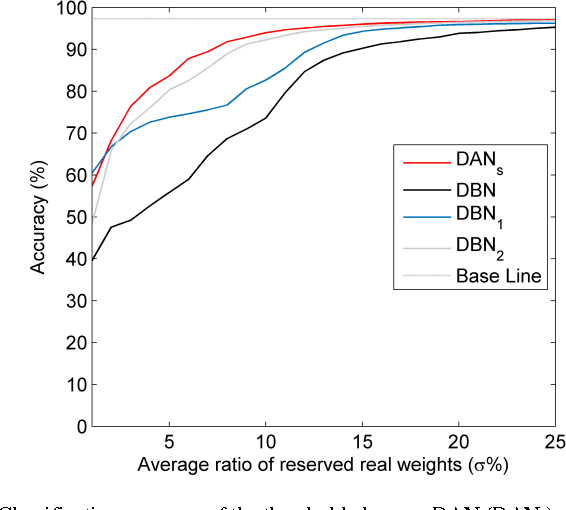
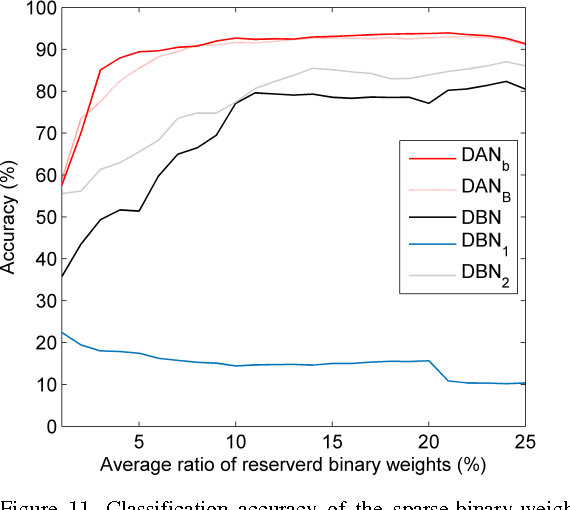
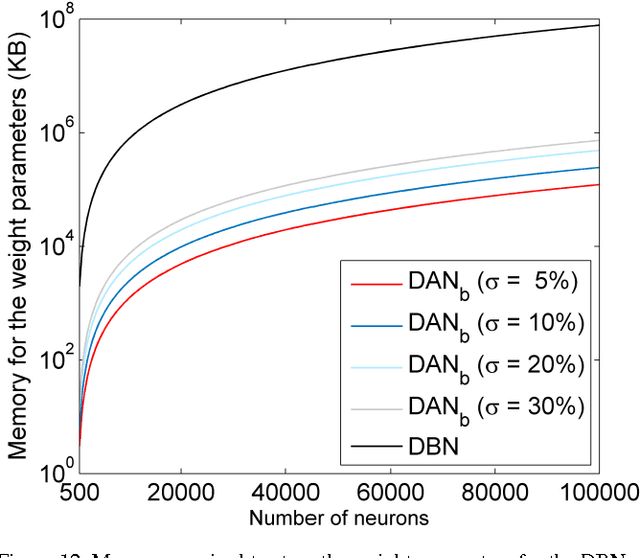
Abstract:Deep neural networks are state-of-the-art models for understanding the content of images, video and raw input data. However, implementing a deep neural network in embedded systems is a challenging task, because a typical deep neural network, such as a Deep Belief Network using 128x128 images as input, could exhaust Giga bytes of memory and result in bandwidth and computing bottleneck. To address this challenge, this paper presents a hardware-oriented deep learning algorithm, named as the Deep Adaptive Network, which attempts to exploit the sparsity in the neural connections. The proposed method adaptively reduces the weights associated with negligible features to zero, leading to sparse feedforward network architecture. Furthermore, since the small proportion of important weights are significantly larger than zero, they can be robustly thresholded and represented using single-bit integers (-1 and +1), leading to implementations of deep neural networks with sparse and binary connections. Our experiments showed that, for the application of recognizing MNIST handwritten digits, the features extracted by a two-layer Deep Adaptive Network with about 25% reserved important connections achieved 97.2% classification accuracy, which was almost the same with the standard Deep Belief Network (97.3%). Furthermore, for efficient hardware implementations, the sparse-and-binary-weighted deep neural network could save about 99.3% memory and 99.9% computation units without significant loss of classification accuracy for pattern recognition applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge