Meng Yang
ResAgent: Entropy-based Prior Point Discovery and Visual Reasoning for Referring Expression Segmentation
Jan 23, 2026Abstract:Referring Expression Segmentation (RES) is a core vision-language segmentation task that enables pixel-level understanding of targets via free-form linguistic expressions, supporting critical applications such as human-robot interaction and augmented reality. Despite the progress of Multimodal Large Language Model (MLLM)-based approaches, existing RES methods still suffer from two key limitations: first, the coarse bounding boxes from MLLMs lead to redundant or non-discriminative point prompts; second, the prevalent reliance on textual coordinate reasoning is unreliable, as it fails to distinguish targets from visually similar distractors. To address these issues, we propose \textbf{\model}, a novel RES framework integrating \textbf{E}ntropy-\textbf{B}ased Point \textbf{D}iscovery (\textbf{EBD}) and \textbf{V}ision-\textbf{B}ased \textbf{R}easoning (\textbf{VBR}). Specifically, EBD identifies high-information candidate points by modeling spatial uncertainty within coarse bounding boxes, treating point selection as an information maximization process. VBR verifies point correctness through joint visual-semantic alignment, abandoning text-only coordinate inference for more robust validation. Built on these components, \model implements a coarse-to-fine workflow: bounding box initialization, entropy-guided point discovery, vision-based validation, and mask decoding. Extensive evaluations on four benchmark datasets (RefCOCO, RefCOCO+, RefCOCOg, and ReasonSeg) demonstrate that \model achieves new state-of-the-art performance across all four benchmarks, highlighting its effectiveness in generating accurate and semantically grounded segmentation masks with minimal prompts.
Granular-ball Guided Masking: Structure-aware Data Augmentation
Dec 24, 2025Abstract:Deep learning models have achieved remarkable success in computer vision, but they still rely heavily on large-scale labeled data and tend to overfit when data are limited or distributions shift. Data augmentation, particularly mask-based information dropping, can enhance robustness by forcing models to explore complementary cues; however, existing approaches often lack structural awareness and may discard essential semantics. We propose Granular-ball Guided Masking (GBGM), a structure-aware augmentation strategy guided by Granular-ball Computing (GBC). GBGM adaptively preserves semantically rich, structurally important regions while suppressing redundant areas through a coarse-to-fine hierarchical masking process, producing augmentations that are both representative and discriminative. Extensive experiments on multiple benchmarks demonstrate consistent improvements in classification accuracy and masked image reconstruction, confirming the effectiveness and broad applicability of the proposed method. Simple and model-agnostic, it integrates seamlessly into CNNs and Vision Transformers and provides a new paradigm for structure-aware data augmentation.
DistillMatch: Leveraging Knowledge Distillation from Vision Foundation Model for Multimodal Image Matching
Sep 19, 2025Abstract:Multimodal image matching seeks pixel-level correspondences between images of different modalities, crucial for cross-modal perception, fusion and analysis. However, the significant appearance differences between modalities make this task challenging. Due to the scarcity of high-quality annotated datasets, existing deep learning methods that extract modality-common features for matching perform poorly and lack adaptability to diverse scenarios. Vision Foundation Model (VFM), trained on large-scale data, yields generalizable and robust feature representations adapted to data and tasks of various modalities, including multimodal matching. Thus, we propose DistillMatch, a multimodal image matching method using knowledge distillation from VFM. DistillMatch employs knowledge distillation to build a lightweight student model that extracts high-level semantic features from VFM (including DINOv2 and DINOv3) to assist matching across modalities. To retain modality-specific information, it extracts and injects modality category information into the other modality's features, which enhances the model's understanding of cross-modal correlations. Furthermore, we design V2I-GAN to boost the model's generalization by translating visible to pseudo-infrared images for data augmentation. Experiments show that DistillMatch outperforms existing algorithms on public datasets.
Seed LiveInterpret 2.0: End-to-end Simultaneous Speech-to-speech Translation with Your Voice
Jul 24, 2025



Abstract:Simultaneous Interpretation (SI) represents one of the most daunting frontiers in the translation industry, with product-level automatic systems long plagued by intractable challenges: subpar transcription and translation quality, lack of real-time speech generation, multi-speaker confusion, and translated speech inflation, especially in long-form discourses. In this study, we introduce Seed-LiveInterpret 2.0, an end-to-end SI model that delivers high-fidelity, ultra-low-latency speech-to-speech generation with voice cloning capabilities. As a fully operational product-level solution, Seed-LiveInterpret 2.0 tackles these challenges head-on through our novel duplex speech-to-speech understanding-generating framework. Experimental results demonstrate that through large-scale pretraining and reinforcement learning, the model achieves a significantly better balance between translation accuracy and latency, validated by human interpreters to exceed 70% correctness in complex scenarios. Notably, Seed-LiveInterpret 2.0 outperforms commercial SI solutions by significant margins in translation quality, while slashing the average latency of cloned speech from nearly 10 seconds to a near-real-time 3 seconds, which is around a near 70% reduction that drastically enhances practical usability.
Morpheus: A Neural-driven Animatronic Face with Hybrid Actuation and Diverse Emotion Control
Jul 22, 2025Abstract:Previous animatronic faces struggle to express emotions effectively due to hardware and software limitations. On the hardware side, earlier approaches either use rigid-driven mechanisms, which provide precise control but are difficult to design within constrained spaces, or tendon-driven mechanisms, which are more space-efficient but challenging to control. In contrast, we propose a hybrid actuation approach that combines the best of both worlds. The eyes and mouth-key areas for emotional expression-are controlled using rigid mechanisms for precise movement, while the nose and cheek, which convey subtle facial microexpressions, are driven by strings. This design allows us to build a compact yet versatile hardware platform capable of expressing a wide range of emotions. On the algorithmic side, our method introduces a self-modeling network that maps motor actions to facial landmarks, allowing us to automatically establish the relationship between blendshape coefficients for different facial expressions and the corresponding motor control signals through gradient backpropagation. We then train a neural network to map speech input to corresponding blendshape controls. With our method, we can generate distinct emotional expressions such as happiness, fear, disgust, and anger, from any given sentence, each with nuanced, emotion-specific control signals-a feature that has not been demonstrated in earlier systems. We release the hardware design and code at https://github.com/ZZongzheng0918/Morpheus-Hardware and https://github.com/ZZongzheng0918/Morpheus-Software.
PacGDC: Label-Efficient Generalizable Depth Completion with Projection Ambiguity and Consistency
Jul 10, 2025Abstract:Generalizable depth completion enables the acquisition of dense metric depth maps for unseen environments, offering robust perception capabilities for various downstream tasks. However, training such models typically requires large-scale datasets with metric depth labels, which are often labor-intensive to collect. This paper presents PacGDC, a label-efficient technique that enhances data diversity with minimal annotation effort for generalizable depth completion. PacGDC builds on novel insights into inherent ambiguities and consistencies in object shapes and positions during 2D-to-3D projection, allowing the synthesis of numerous pseudo geometries for the same visual scene. This process greatly broadens available geometries by manipulating scene scales of the corresponding depth maps. To leverage this property, we propose a new data synthesis pipeline that uses multiple depth foundation models as scale manipulators. These models robustly provide pseudo depth labels with varied scene scales, affecting both local objects and global layouts, while ensuring projection consistency that supports generalization. To further diversify geometries, we incorporate interpolation and relocation strategies, as well as unlabeled images, extending the data coverage beyond the individual use of foundation models. Extensive experiments show that PacGDC achieves remarkable generalizability across multiple benchmarks, excelling in diverse scene semantics/scales and depth sparsity/patterns under both zero-shot and few-shot settings. Code: https://github.com/Wang-xjtu/PacGDC.
Real-World Depth Recovery via Structure Uncertainty Modeling and Inaccurate GT Depth Fitting
Apr 16, 2025Abstract:The low-quality structure in raw depth maps is prevalent in real-world RGB-D datasets, which makes real-world depth recovery a critical task in recent years. However, the lack of paired raw-ground truth (raw-GT) data in the real world poses challenges for generalized depth recovery. Existing methods insufficiently consider the diversity of structure misalignment in raw depth maps, which leads to poor generalization in real-world depth recovery. Notably, random structure misalignments are not limited to raw depth data but also affect GT depth in real-world datasets. In the proposed method, we tackle the generalization problem from both input and output perspectives. For input, we enrich the diversity of structure misalignment in raw depth maps by designing a new raw depth generation pipeline, which helps the network avoid overfitting to a specific condition. Furthermore, a structure uncertainty module is designed to explicitly identify the misaligned structure for input raw depth maps to better generalize in unseen scenarios. Notably the well-trained depth foundation model (DFM) can help the structure uncertainty module estimate the structure uncertainty better. For output, a robust feature alignment module is designed to precisely align with the accurate structure of RGB images avoiding the interference of inaccurate GT depth. Extensive experiments on multiple datasets demonstrate the proposed method achieves competitive accuracy and generalization capabilities across various challenging raw depth maps.
Multimodal Image Matching based on Frequency-domain Information of Local Energy Response
Mar 26, 2025Abstract:Complicated nonlinear intensity differences, nonlinear local geometric distortions, noises and rotation transformation are main challenges in multimodal image matching. In order to solve these problems, we propose a method based on Frequency-domain Information of Local Energy Response called FILER. The core of FILER is the local energy response model based on frequency-domain information, which can overcome the effect of nonlinear intensity differences. To improve the robustness to local nonlinear geometric distortions and noises, we design a new edge structure enhanced feature detector and convolutional feature weighted descriptor, respectively. In addition, FILER overcomes the sensitivity of the frequency-domain information to the rotation angle and achieves rotation invariance. Extensive experiments multimodal image pairs show that FILER outperforms other state-of-the-art algorithms and has good robustness and universality.
A Causality-Inspired Model for Intima-Media Thickening Assessment in Ultrasound Videos
Mar 16, 2025Abstract:Carotid atherosclerosis represents a significant health risk, with its early diagnosis primarily dependent on ultrasound-based assessments of carotid intima-media thickening. However, during carotid ultrasound screening, significant view variations cause style shifts, impairing content cues related to thickening, such as lumen anatomy, which introduces spurious correlations that hinder assessment. Therefore, we propose a novel causal-inspired method for assessing carotid intima-media thickening in frame-wise ultrasound videos, which focuses on two aspects: eliminating spurious correlations caused by style and enhancing causal content correlations. Specifically, we introduce a novel Spurious Correlation Elimination (SCE) module to remove non-causal style effects by enforcing prediction invariance with style perturbations. Simultaneously, we propose a Causal Equivalence Consolidation (CEC) module to strengthen causal content correlation through adversarial optimization during content randomization. Simultaneously, we design a Causal Transition Augmentation (CTA) module to ensure smooth causal flow by integrating an auxiliary pathway with text prompts and connecting it through contrastive learning. The experimental results on our in-house carotid ultrasound video dataset achieved an accuracy of 86.93\%, demonstrating the superior performance of the proposed method. Code is available at \href{https://github.com/xielaobanyy/causal-imt}{https://github.com/xielaobanyy/causal-imt}.
PUGS: Zero-shot Physical Understanding with Gaussian Splatting
Feb 17, 2025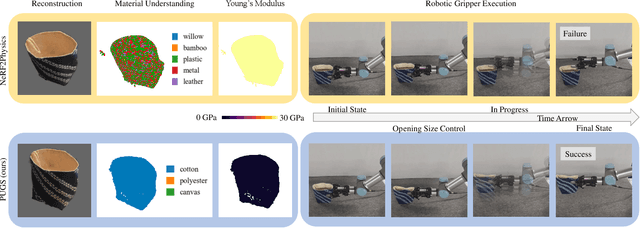
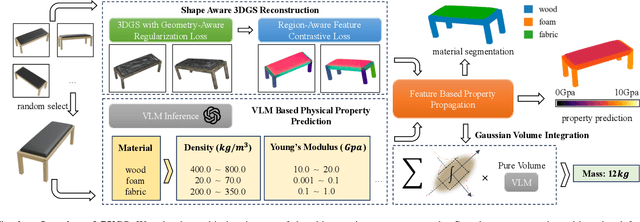

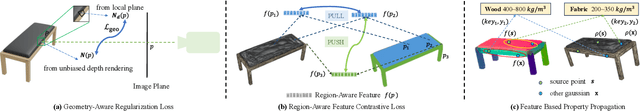
Abstract:Current robotic systems can understand the categories and poses of objects well. But understanding physical properties like mass, friction, and hardness, in the wild, remains challenging. We propose a new method that reconstructs 3D objects using the Gaussian splatting representation and predicts various physical properties in a zero-shot manner. We propose two techniques during the reconstruction phase: a geometry-aware regularization loss function to improve the shape quality and a region-aware feature contrastive loss function to promote region affinity. Two other new techniques are designed during inference: a feature-based property propagation module and a volume integration module tailored for the Gaussian representation. Our framework is named as zero-shot physical understanding with Gaussian splatting, or PUGS. PUGS achieves new state-of-the-art results on the standard benchmark of ABO-500 mass prediction. We provide extensive quantitative ablations and qualitative visualization to demonstrate the mechanism of our designs. We show the proposed methodology can help address challenging real-world grasping tasks. Our codes, data, and models are available at https://github.com/EverNorif/PUGS
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge