Yueyang Teng
Progressive $\mathcal{J}$-Invariant Self-supervised Learning for Low-Dose CT Denoising
Jan 28, 2026Abstract:Self-supervised learning has been increasingly investigated for low-dose computed tomography (LDCT) image denoising, as it alleviates the dependence on paired normal-dose CT (NDCT) data, which are often difficult to collect. However, many existing self-supervised blind-spot denoising methods suffer from training inefficiencies and suboptimal performance due to restricted receptive fields. To mitigate this issue, we propose a novel Progressive $\mathcal{J}$-invariant Learning that maximizes the use of $\mathcal{J}$-invariant to enhance LDCT denoising performance. We introduce a step-wise blind-spot denoising mechanism that enforces conditional independence in a progressive manner, enabling more fine-grained learning for denoising. Furthermore, we explicitly inject a combination of controlled Gaussian and Poisson noise during training to regularize the denoising process and mitigate overfitting. Extensive experiments on the Mayo LDCT dataset demonstrate that the proposed method consistently outperforms existing self-supervised approaches and achieves performance comparable to, or better than, several representative supervised denoising methods.
Progressive self-supervised blind-spot denoising method for LDCT denoising
Jan 20, 2026Abstract:Self-supervised learning is increasingly investigated for low-dose computed tomography (LDCT) image denoising, as it alleviates the dependence on paired normal-dose CT (NDCT) data, which are often difficult to acquire in clinical practice. In this paper, we propose a novel self-supervised training strategy that relies exclusively on LDCT images. We introduce a step-wise blind-spot denoising mechanism that enforces conditional independence in a progressive manner, enabling more fine-grained denoising learning. In addition, we add Gaussian noise to LDCT images, which acts as a regularization and mitigates overfitting. Extensive experiments on the Mayo LDCT dataset demonstrate that the proposed method consistently outperforms existing self-supervised approaches and achieves performance comparable to, or better than, several representative supervised denoising methods.
Self-supervised Noise2noise Method Utilizing Corrupted Images with a Modular Network for LDCT Denoising
Aug 13, 2023Abstract:Deep learning is a very promising technique for low-dose computed tomography (LDCT) image denoising. However, traditional deep learning methods require paired noisy and clean datasets, which are often difficult to obtain. This paper proposes a new method for performing LDCT image denoising with only LDCT data, which means that normal-dose CT (NDCT) is not needed. We adopt a combination including the self-supervised noise2noise model and the noisy-as-clean strategy. First, we add a second yet similar type of noise to LDCT images multiple times. Note that we use LDCT images based on the noisy-as-clean strategy for corruption instead of NDCT images. Then, the noise2noise model is executed with only the secondary corrupted images for training. We select a modular U-Net structure from several candidates with shared parameters to perform the task, which increases the receptive field without increasing the parameter size. The experimental results obtained on the Mayo LDCT dataset show the effectiveness of the proposed method compared with that of state-of-the-art deep learning methods. The developed code is available at https://github.com/XYuan01/Self-supervised-Noise2Noise-for-LDCT.
Ultrasonic Image's Annotation Removal: A Self-supervised Noise2Noise Approach
Jul 09, 2023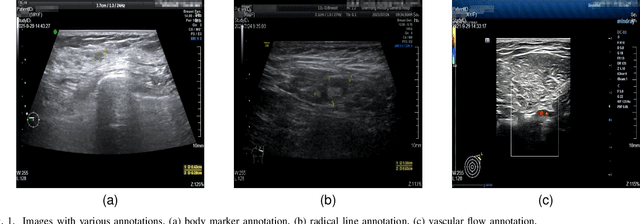
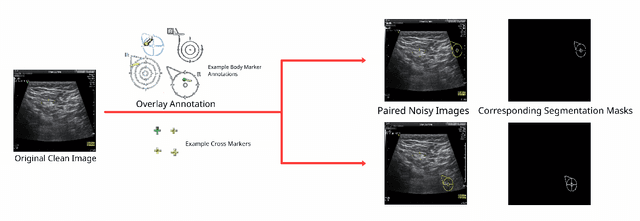
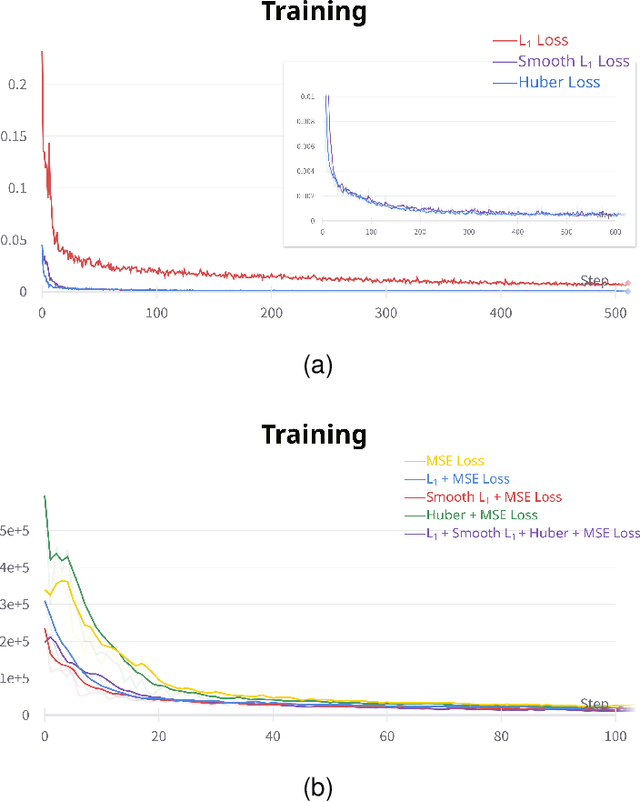
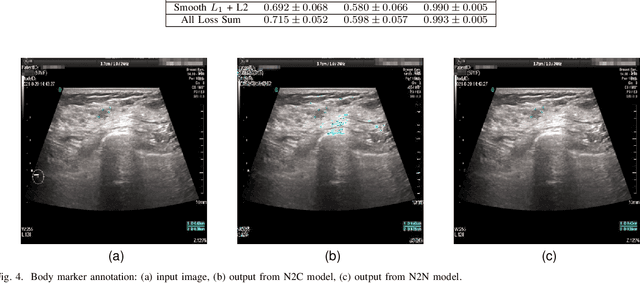
Abstract:Accurately annotated ultrasonic images are vital components of a high-quality medical report. Hospitals often have strict guidelines on the types of annotations that should appear on imaging results. However, manually inspecting these images can be a cumbersome task. While a neural network could potentially automate the process, training such a model typically requires a dataset of paired input and target images, which in turn involves significant human labour. This study introduces an automated approach for detecting annotations in images. This is achieved by treating the annotations as noise, creating a self-supervised pretext task and using a model trained under the Noise2Noise scheme to restore the image to a clean state. We tested a variety of model structures on the denoising task against different types of annotation, including body marker annotation, radial line annotation, etc. Our results demonstrate that most models trained under the Noise2Noise scheme outperformed their counterparts trained with noisy-clean data pairs. The costumed U-Net yielded the most optimal outcome on the body marker annotation dataset, with high scores on segmentation precision and reconstruction similarity. We released our code at https://github.com/GrandArth/UltrasonicImage-N2N-Approach.
NL-CS Net: Deep Learning with Non-Local Prior for Image Compressive Sensing
May 06, 2023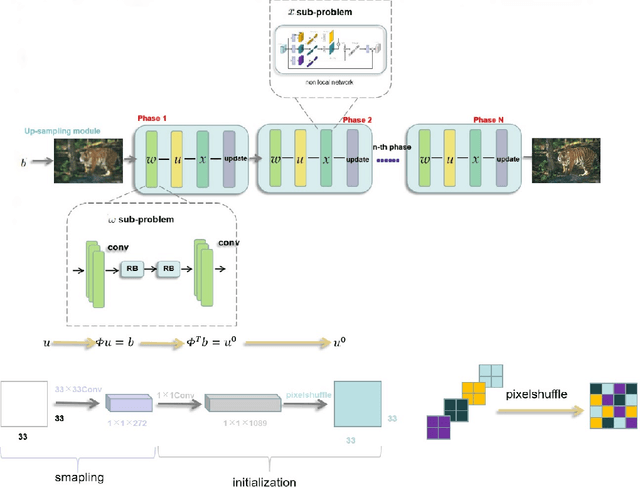
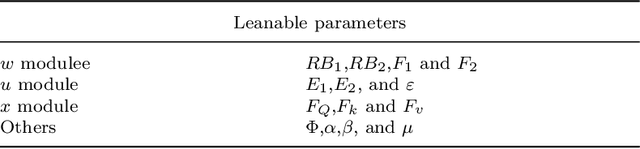
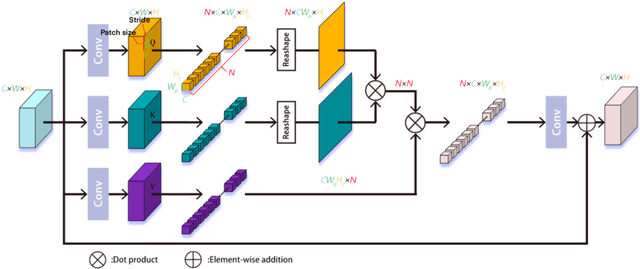
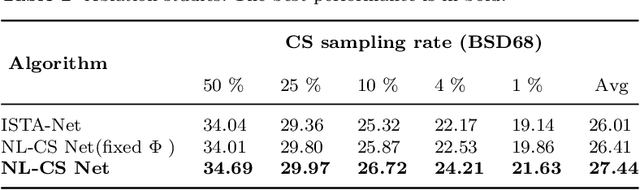
Abstract:Deep learning has been applied to compressive sensing (CS) of images successfully in recent years. However, existing network-based methods are often trained as the black box, in which the lack of prior knowledge is often the bottleneck for further performance improvement. To overcome this drawback, this paper proposes a novel CS method using non-local prior which combines the interpretability of the traditional optimization methods with the speed of network-based methods, called NL-CS Net. We unroll each phase from iteration of the augmented Lagrangian method solving non-local and sparse regularized optimization problem by a network. NL-CS Net is composed of the up-sampling module and the recovery module. In the up-sampling module, we use learnable up-sampling matrix instead of a predefined one. In the recovery module, patch-wise non-local network is employed to capture long-range feature correspondences. Important parameters involved (e.g. sampling matrix, nonlinear transforms, shrinkage thresholds, step size, $etc.$) are learned end-to-end, rather than hand-crafted. Furthermore, to facilitate practical implementation, orthogonal and binary constraints on the sampling matrix are simultaneously adopted. Extensive experiments on natural images and magnetic resonance imaging (MRI) demonstrate that the proposed method outperforms the state-of-the-art methods while maintaining great interpretability and speed.
3D PETCT Tumor Lesion Segmentation via GCN Refinement
Feb 24, 2023Abstract:Whole-body PET/CT scan is an important tool for diagnosing various malignancies (e.g., malignant melanoma, lymphoma, or lung cancer), and accurate segmentation of tumors is a key part for subsequent treatment. In recent years, CNN-based segmentation methods have been extensively investigated. However, these methods often give inaccurate segmentation results, such as over-segmentation and under-segmentation. Therefore, to address such issues, we propose a post-processing method based on a graph convolutional neural network (GCN) to refine inaccurate segmentation parts and improve the overall segmentation accuracy. Firstly, nnUNet is used as an initial segmentation framework, and the uncertainty in the segmentation results is analyzed. Certainty and uncertainty nodes establish the nodes of a graph neural network. Each node and its 6 neighbors form an edge, and 32 nodes are randomly selected for uncertain nodes to form edges. The highly uncertain nodes are taken as the subsequent refinement targets. Secondly, the nnUNet result of the certainty nodes is used as label to form a semi-supervised graph network problem, and the uncertainty part is optimized through training the GCN network to improve the segmentation performance. This describes our proposed nnUNet-GCN segmentation framework. We perform tumor segmentation experiments on the PET/CT dataset in the MICCIA2022 autoPET challenge. Among them, 30 cases are randomly selected for testing, and the experimental results show that the false positive rate is effectively reduced with nnUNet-GCN refinement. In quantitative analysis, there is an improvement of 2.12 % on the average Dice score, 6.34 on 95 % Hausdorff Distance (HD95), and 1.72 on average symmetric surface distance (ASSD). The quantitative and qualitative evaluation results show that GCN post-processing methods can effectively improve tumor segmentation performance.
QS-ADN: Quasi-Supervised Artifact Disentanglement Network for Low-Dose CT Image Denoising by Local Similarity Among Unpaired Data
Feb 08, 2023Abstract:Deep learning has been successfully applied to low-dose CT (LDCT) image denoising for reducing potential radiation risk. However, the widely reported supervised LDCT denoising networks require a training set of paired images, which is expensive to obtain and cannot be perfectly simulated. Unsupervised learning utilizes unpaired data and is highly desirable for LDCT denoising. As an example, an artifact disentanglement network (ADN) relies on unparied images and obviates the need for supervision but the results of artifact reduction are not as good as those through supervised learning.An important observation is that there is often hidden similarity among unpaired data that can be utilized. This paper introduces a new learning mode, called quasi-supervised learning, to empower the ADN for LDCT image denoising.For every LDCT image, the best matched image is first found from an unpaired normal-dose CT (NDCT) dataset. Then, the matched pairs and the corresponding matching degree as prior information are used to construct and train our ADN-type network for LDCT denoising.The proposed method is different from (but compatible with) supervised and semi-supervised learning modes and can be easily implemented by modifying existing networks. The experimental results show that the method is competitive with state-of-the-art methods in terms of noise suppression and contextual fidelity. The code and working dataset are publicly available at https://github.com/ruanyuhui/ADN-QSDL.git.
EBHI-Seg: A Novel Enteroscope Biopsy Histopathological Haematoxylin and Eosin Image Dataset for Image Segmentation Tasks
Dec 06, 2022



Abstract:Background and Purpose: Colorectal cancer is a common fatal malignancy, the fourth most common cancer in men, and the third most common cancer in women worldwide. Timely detection of cancer in its early stages is essential for treating the disease. Currently, there is a lack of datasets for histopathological image segmentation of rectal cancer, which often hampers the assessment accuracy when computer technology is used to aid in diagnosis. Methods: This present study provided a new publicly available Enteroscope Biopsy Histopathological Hematoxylin and Eosin Image Dataset for Image Segmentation Tasks (EBHI-Seg). To demonstrate the validity and extensiveness of EBHI-Seg, the experimental results for EBHI-Seg are evaluated using classical machine learning methods and deep learning methods. Results: The experimental results showed that deep learning methods had a better image segmentation performance when utilizing EBHI-Seg. The maximum accuracy of the Dice evaluation metric for the classical machine learning method is 0.948, while the Dice evaluation metric for the deep learning method is 0.965. Conclusion: This publicly available dataset contained 5,170 images of six types of tumor differentiation stages and the corresponding ground truth images. The dataset can provide researchers with new segmentation algorithms for medical diagnosis of colorectal cancer, which can be used in the clinical setting to help doctors and patients.
Quasi-supervised Learning for Super-resolution PET
Sep 03, 2022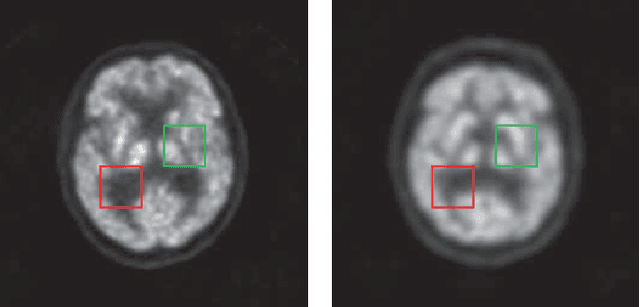
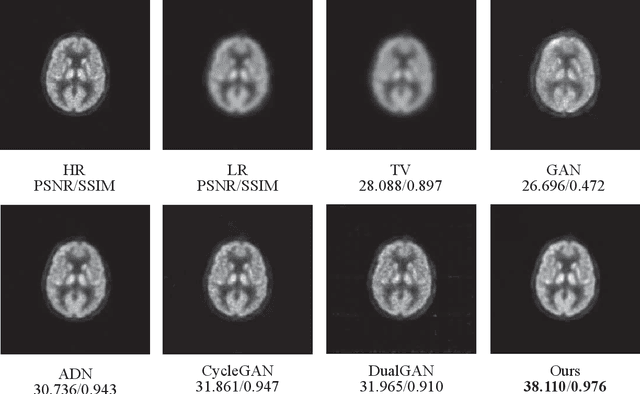
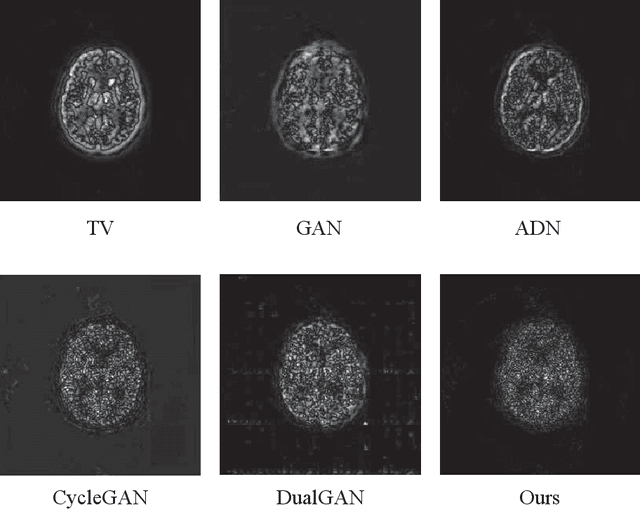
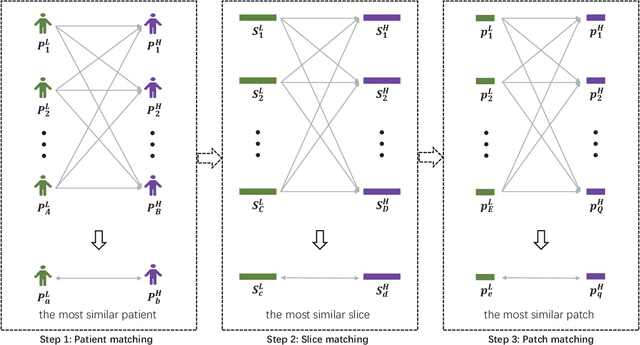
Abstract:Low resolution of positron emission tomography (PET) limits its diagnostic performance. Deep learning has been successfully applied to achieve super-resolution PET. However, commonly used supervised learning methods in this context require many pairs of low- and high-resolution (LR and HR) PET images. Although unsupervised learning utilizes unpaired images, the results are not as good as that obtained with supervised deep learning. In this paper, we propose a quasi-supervised learning method, which is a new type of weakly-supervised learning methods, to recover HR PET images from LR counterparts by leveraging similarity between unpaired LR and HR image patches. Specifically, LR image patches are taken from a patient as inputs, while the most similar HR patches from other patients are found as labels. The similarity between the matched HR and LR patches serves as a prior for network construction. Our proposed method can be implemented by designing a new network or modifying an existing network. As an example in this study, we have modified the cycle-consistent generative adversarial network (CycleGAN) for super-resolution PET. Our numerical and experimental results qualitatively and quantitatively show the merits of our method relative to the state-ofthe-art methods. The code is publicly available at https://github.com/PigYang-ops/CycleGAN-QSDL.
Adaptive Weighted Nonnegative Matrix Factorization for Robust Feature Representation
Jun 07, 2022
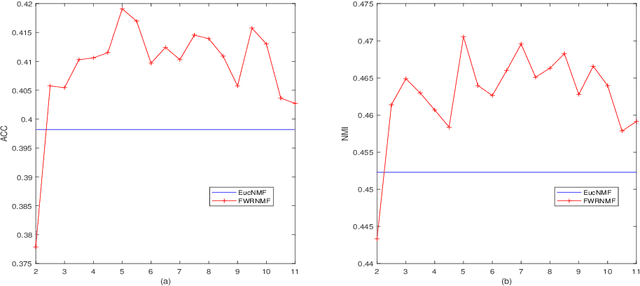
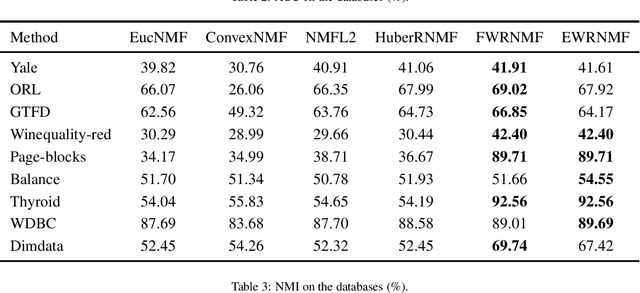
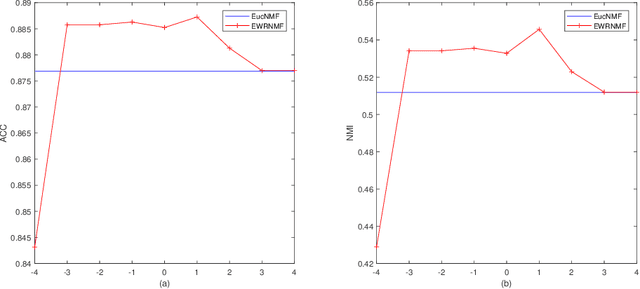
Abstract:Nonnegative matrix factorization (NMF) has been widely used to dimensionality reduction in machine learning. However, the traditional NMF does not properly handle outliers, so that it is sensitive to noise. In order to improve the robustness of NMF, this paper proposes an adaptive weighted NMF, which introduces weights to emphasize the different importance of each data point, thus the algorithmic sensitivity to noisy data is decreased. It is very different from the existing robust NMFs that use a slow growth similarity measure. Specifically, two strategies are proposed to achieve this: fuzzier weighted technique and entropy weighted regularized technique, and both of them lead to an iterative solution with a simple form. Experimental results showed that new methods have more robust feature representation on several real datasets with noise than exsiting methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge